Improving counterfactual reasoning with kernelised dynamic mixing models
- PMID: 30419029
- PMCID: PMC6231902
- DOI: 10.1371/journal.pone.0205839
Improving counterfactual reasoning with kernelised dynamic mixing models
Abstract
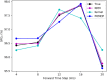
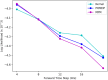
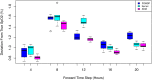
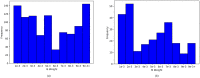
Simulation-based approaches to disease progression allow us to make counterfactual predictions about the effects of an untried series of treatment choices. However, building accurate simulators of disease progression is challenging, limiting the utility of these approaches for real world treatment planning. In this work, we present a novel simulation-based reinforcement learning approach that mixes between models and kernel-based approaches to make its forward predictions. On two real world tasks, managing sepsis and treating HIV, we demonstrate that our approach both learns state-of-the-art treatment policies and can make accurate forward predictions about the effects of treatments on unseen patients.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
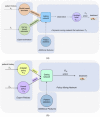

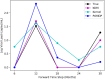
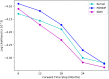
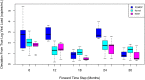
References
-
- Che Z, Kale D, Li W, Bahadori MT, Liu Y. Deep computational phenotyping. In: KDD; 2015.
-
- Bogojeska J, Stöckel D, Zazzi M, Kaiser R, Incardona F, Rosen-Zvi M, et al. History-alignment models for bias-aware prediction of virological response to HIV combination therapy. In: AISTATS; 2012. p. 118–126.

