Rapidly evolving changes and gene loss associated with host switching in Corynebacterium pseudotuberculosis
- PMID: 30419061
- PMCID: PMC6231662
- DOI: 10.1371/journal.pone.0207304
Rapidly evolving changes and gene loss associated with host switching in Corynebacterium pseudotuberculosis
Abstract
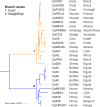
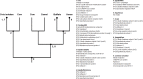
Phylogenomics and genome scale positive selection analyses were performed on 29 Corynebacterium pseudotuberculosis genomes that were isolated from different hosts, including representatives of the Ovis and Equi biovars. A total of 27 genes were identified as undergoing adaptive changes. An analysis of the clades within this species and these biovars, the genes specific to each branch, and the genes responding to selective pressure show clear differences, indicating that adaptation and specialization is occurring in different clades. These changes are often correlated with the isolation host but could indicate responses to some undetermined factor in the respective niches. The fact that some of these more-rapidly evolving genes have homology to known virulence factors, antimicrobial resistance genes and drug targets shows that this type of analysis could be used to identify novel targets, and that these could be used as a way to control this pathogen.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
A shift in the virulence potential of Corynebacterium pseudotuberculosis biovar ovis after passage in a murine host demonstrated through comparative proteomics.BMC Microbiol. 2017 Mar 22;17(1):55. doi: 10.1186/s12866-017-0925-6. BMC Microbiol. 2017. PMID: 28327085 Free PMC article.
-
A comparative pan-genomic analysis of 53 C. pseudotuberculosis strains based on functional domains.J Biomol Struct Dyn. 2021 Nov;39(18):6974-6986. doi: 10.1080/07391102.2020.1805017. Epub 2020 Aug 11. J Biomol Struct Dyn. 2021. PMID: 32779519
-
Transcriptome analysis of Corynebacterium pseudotuberculosis biovar Equi in two conditions of the environmental stress.Gene. 2018 Nov 30;677:349-360. doi: 10.1016/j.gene.2018.08.028. Epub 2018 Aug 8. Gene. 2018. PMID: 30098432
-
[PATHOGENICITY FACTORS OF CORYNEBACTERIUM NON DIPHTHERIAE].Zh Mikrobiol Epidemiol Immunobiol. 2016 May;(3):97-104. Zh Mikrobiol Epidemiol Immunobiol. 2016. PMID: 30695460 Review. Russian.
-
A description of genes of Corynebacterium pseudotuberculosis useful in diagnostics and vaccine applications.Genet Mol Res. 2008 Mar 18;7(1):252-60. doi: 10.4238/vol7-1gmr438. Genet Mol Res. 2008. PMID: 18551390 Review.
Cited by
-
Corynebacterium pseudotuberculosis: Whole genome sequencing reveals unforeseen and relevant genetic diversity in this pathogen.PLoS One. 2024 Aug 26;19(8):e0309282. doi: 10.1371/journal.pone.0309282. eCollection 2024. PLoS One. 2024. PMID: 39186721 Free PMC article.
-
Comprehensive genome analysis and comparisons of the swine pathogen, Chlamydia suis reveals unique ORFs and candidate host-specificity factors.Pathog Dis. 2021 Mar 10;79(2):ftaa035. doi: 10.1093/femspd/ftaa035. Pathog Dis. 2021. PMID: 32639528 Free PMC article.
-
Ovine and Caprine Strains of Corynebacterium pseudotuberculosis on Czech Farms-A Comparative Study.Microorganisms. 2024 Apr 27;12(5):875. doi: 10.3390/microorganisms12050875. Microorganisms. 2024. PMID: 38792705 Free PMC article.
-
Insights into the Metabolism and Evolution of the Genus Acidiphilium, a Typical Acidophile in Acid Mine Drainage.mSystems. 2020 Nov 17;5(6):e00867-20. doi: 10.1128/mSystems.00867-20. mSystems. 2020. PMID: 33203689 Free PMC article.
-
Resequencing and characterization of the first Corynebacterium pseudotuberculosis genome isolated from camel.PeerJ. 2024 Jan 30;12:e16513. doi: 10.7717/peerj.16513. eCollection 2024. PeerJ. 2024. PMID: 38313017 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

