Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure
- PMID: 30419824
- PMCID: PMC6233272
- DOI: 10.1186/s12864-018-5213-9
Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure
Abstract
Background: Current heart failure (HF) treatment is based on targeting symptoms and left ventricle dysfunction severity, relying on a common HF pathway paradigm to justify common treatments for HF patients. This common strategy may belie an incomplete understanding of heterogeneous underlying mechanisms and could be a barrier to more precise treatments. We hypothesized we could use RNA-sequencing (RNA-seq) in human heart tissue to delineate HF etiology-specific gene expression signatures.
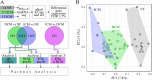
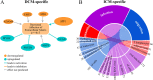
Results: RNA-seq from 64 human left ventricular samples: 37 dilated (DCM), 13 ischemic (ICM), and 14 non-failing (NF). Using a multi-analytic approach including covariate adjustment for age and sex, differentially expressed genes (DEGs) were identified characterizing HF and disease-specific expression. Pathway analysis investigated enrichment for biologically relevant pathways and functions. DCM vs NF and ICM vs NF had shared HF-DEGs that were enriched for the fetal gene program and mitochondrial dysfunction. DCM-specific DEGs were enriched for cell-cell and cell-matrix adhesion pathways. ICM-specific DEGs were enriched for cytoskeletal and immune pathway activation. Using the ICM and DCM DEG signatures from our data we were able to correctly classify the phenotypes of 24/31 ICM and 32/36 DCM samples from publicly available replication datasets.
Conclusions: Our results demonstrate the commonality of mitochondrial dysfunction in end-stage HF but more importantly reveal key etiology-specific signatures. Dysfunctional cell-cell and cell-matrix adhesion signatures typified DCM whereas signals related to immune and fibrotic responses were seen in ICM. These findings suggest that transcriptome signatures may distinguish end-stage heart failure, shedding light on underlying biological differences between ICM and DCM.
Keywords: Cardiomyopathy; Gene expression; Heart failure; RNA-seq; Transcriptome.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the Colorado Multiple Institutional Review Board (COMIRB, protocol 01-568) whereby transplant-listed patients signed written consent for use of their explanted hearts for research purposes. NF hearts were obtained by the local organ procurement agency whereby family members of organ donors signed written consent for research use of explanted cardiac tissue.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
-
- Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Jr, Drazner MH, et al. 2013 ACCF/AHA guideline for the management of heart failure: executive summary: a report of the American College of Cardiology Foundation/American Heart Association task force on practice guidelines. Circulation. 2013;128(16):1810–1852. doi: 10.1161/CIR.0b013e31829e8807. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

