Human fluids alter DNA-acquisition in Acinetobacter baumannii
- PMID: 30420211
- PMCID: PMC6372348
- DOI: 10.1016/j.diagmicrobio.2018.10.010
Human fluids alter DNA-acquisition in Acinetobacter baumannii
Abstract
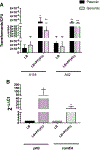
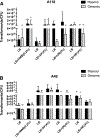
Transformation is one of the mechanisms of acquisition of foreign genetic material leading to the emergence of multidrug resistant (MDR) bacteria. Recently, human serum albumin (HSA) was shown to specifically increase transformation frequency in the nosocomial pathogen Acinetobacter baumannii. To further assess the relevance of HSA as a possible modulator of A. baumannii transformation in host-pathogen interactions, in this work we examined the effect of different human fluids. We observed a significant increase in transformation frequencies in the presence of pleural fluid, whole blood cells and liquid ascites, and to a lesser extent with urine. The observed effects correlate with both HSA and bacterial content found in the assayed patient fluids. Taken together, these results are in agreement with our previous findings that highlight HSA as a possible host signal with the ability to trigger natural transformation in A. baumannii.
Keywords: Acinetobacter baumannii; Antibiotic resistance; DNA-acquisition; Human serum albumin; Pleural fluid; Transformation.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Serum Albumin and Ca2+ Are Natural Competence Inducers in the Human Pathogen Acinetobacter baumannii.Antimicrob Agents Chemother. 2016 Jul 22;60(8):4920-9. doi: 10.1128/AAC.00529-16. Print 2016 Aug. Antimicrob Agents Chemother. 2016. PMID: 27270286 Free PMC article.
-
Fluorescence-Based Detection of Natural Transformation in Drug-Resistant Acinetobacter baumannii.J Bacteriol. 2018 Sep 10;200(19):e00181-18. doi: 10.1128/JB.00181-18. Print 2018 Oct 1. J Bacteriol. 2018. PMID: 30012729 Free PMC article.
-
Effect of Host Human Products on Natural Transformation in Acinetobacter baumannii.Curr Microbiol. 2019 Aug;76(8):950-953. doi: 10.1007/s00284-017-1417-5. Epub 2018 Jan 13. Curr Microbiol. 2019. PMID: 29332139 Free PMC article.
-
Mechanisms Protecting Acinetobacter baumannii against Multiple Stresses Triggered by the Host Immune Response, Antibiotics and Outside-Host Environment.Int J Mol Sci. 2020 Jul 31;21(15):5498. doi: 10.3390/ijms21155498. Int J Mol Sci. 2020. PMID: 32752093 Free PMC article. Review.
-
Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options.Int J Antimicrob Agents. 2012 Feb;39(2):105-14. doi: 10.1016/j.ijantimicag.2011.10.004. Epub 2011 Nov 22. Int J Antimicrob Agents. 2012. PMID: 22113193 Review.
Cited by
-
Cerebrospinal fluid (CSF) augments metabolism and virulence expression factors in Acinetobacter baumannii.Sci Rep. 2021 Feb 26;11(1):4737. doi: 10.1038/s41598-021-81714-6. Sci Rep. 2021. PMID: 33637791 Free PMC article.
-
DNA uptake and twitching motility are controlled by the small RNA Arp through repression of pilin translation in Acinetobacter baumannii.bioRxiv [Preprint]. 2025 Jul 19:2025.07.19.665661. doi: 10.1101/2025.07.19.665661. bioRxiv. 2025. PMID: 40791418 Free PMC article. Preprint.
-
Acinetobacter baumannii, a Multidrug-Resistant Opportunistic Pathogen in New Habitats: A Systematic Review.Microorganisms. 2024 Mar 23;12(4):644. doi: 10.3390/microorganisms12040644. Microorganisms. 2024. PMID: 38674589 Free PMC article. Review.
-
Human serum albumin (HSA) regulates the expression of histone-like nucleoid structure protein (H-NS) in Acinetobacter baumannii.Sci Rep. 2022 Aug 27;12(1):14644. doi: 10.1038/s41598-022-19012-y. Sci Rep. 2022. PMID: 36030268 Free PMC article.
-
Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress.Sci Rep. 2019 Nov 21;9(1):17251. doi: 10.1038/s41598-019-53847-2. Sci Rep. 2019. PMID: 31754169 Free PMC article.
References
-
- WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics News release by the WHO. 2017.
-
- Gerischer U, Ornston LN. Dependence of linkage of alleles on their physical distance in natural transformation of Acinetobacter sp. strain ADP1. Arch Microbiol 2001;176:465–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

