Tri-arginine exosite patch of caspase-6 recruits substrates for hydrolysis
- PMID: 30420425
- PMCID: PMC6322879
- DOI: 10.1074/jbc.RA118.005914
Tri-arginine exosite patch of caspase-6 recruits substrates for hydrolysis
Abstract
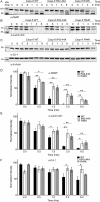
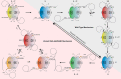
Caspases are cysteine-aspartic proteases involved in the regulation of programmed cell death (apoptosis) and a number of other biological processes. Despite overall similarities in structure and active-site composition, caspases show striking selectivity for particular protein substrates. Exosites are emerging as one of the mechanisms by which caspases can recruit, engage, and orient these substrates for proper hydrolysis. Following computational analyses and database searches for candidate exosites, we utilized site-directed mutagenesis to identify a new exosite in caspase-6 at the hinge between the disordered N-terminal domain (NTD), residues 23-45, and core of the caspase-6 structure. We observed that substitutions of the tri-arginine patch Arg-42-Arg-44 or the R44K cancer-associated mutation in caspase-6 markedly alter its rates of protein substrate hydrolysis. Notably, turnover of protein substrates but not of short peptide substrates was affected by these exosite alterations, underscoring the importance of this region for protein substrate recruitment. Hydrogen-deuterium exchange MS-mediated interrogation of the intrinsic dynamics of these enzymes suggested the presence of a substrate-binding platform encompassed by the NTD and the 240's region (containing residues 236-246), which serves as a general exosite for caspase-6-specific substrate recruitment. In summary, we have identified an exosite on caspase-6 that is critical for protein substrate recognition and turnover and therefore highly relevant for diseases such as cancer in which caspase-6-mediated apoptosis is often disrupted, and in neurodegeneration in which caspase-6 plays a central role.
Keywords: allosteric regulation; apoptosis; cancer; caspase-6; cysteine protease; hydrogen–deuterium exchange; mass spectrometry; neurodegeneration; protein dynamic; substrate recognition; substrate specificity; tri-arginine exosite.
© 2019 MacPherson et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures







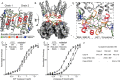
References
-
- Greenblatt M. S., Bennett W. P., Hollstein M., and Harris C. C. (1994) Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 54, 4855–4878 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

