Brain injury induces HIF-1α-dependent transcriptional activation of LRRK2 that exacerbates brain damage
- PMID: 30420654
- PMCID: PMC6232134
- DOI: 10.1038/s41419-018-1180-y
Brain injury induces HIF-1α-dependent transcriptional activation of LRRK2 that exacerbates brain damage
Abstract
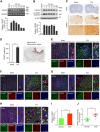
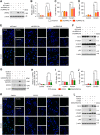
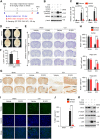
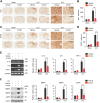
Leucine-rich repeat kinase 2 (LRRK2), originally identified as a causative genetic factor in Parkinson's disease, is now associated with a number of pathologies. Here, we show that brain injury induces a robust expression of endogenous LRRK2 and suggest a role of LRRK2 after injury. We found that various in vitro and in vivo models of traumatic brain injury (TBI) markedly enhanced LRRK2 expression in neurons and also increased the level of hypoxia-inducible factor (HIF)-1α. Luciferase reporter assay and chromatin immunoprecipitation revealed direct binding of HIF-1α in LRRK2 proximal promoter. We also found that HIF-1α-dependent transcriptional induction of LRRK2 exacerbated neuronal cell death following injury. Furthermore, application of G1023, a specific, brain-permeable inhibitor of LRRK2, substantially prevented brain tissue damage, cell death, and inflammatory response and alleviated motor and cognitive defects induced by controlled cortical impact injury. Together, these results suggest HIF-1α-LRRK2 axis as a potential therapeutic target for brain injury.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Gilks WP, et al. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet. 2005;365:415–416. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

