Rescue of recurrent deep intronic mutation underlying cell type-dependent quantitative NEMO deficiency
- PMID: 30422821
- PMCID: PMC6355244
- DOI: 10.1172/JCI124011
Rescue of recurrent deep intronic mutation underlying cell type-dependent quantitative NEMO deficiency
Abstract
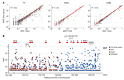
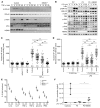
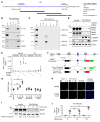
X-linked dominant incontinentia pigmenti (IP) and X-linked recessive anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) are caused by loss-of-function and hypomorphic IKBKG (also known as NEMO) mutations, respectively. We describe a European mother with mild IP and a Japanese mother without IP, whose 3 boys with EDA-ID died from ID. We identify the same private variant in an intron of IKBKG, IVS4+866 C>T, which was inherited from and occurred de novo in the European mother and Japanese mother, respectively. This mutation creates a new splicing donor site, giving rise to a 44-nucleotide pseudoexon (PE) generating a frameshift. Its leakiness accounts for NF-κB activation being impaired but not abolished in the boys' cells. However, aberrant splicing rates differ between cell types, with WT NEMO mRNA and protein levels ranging from barely detectable in leukocytes to residual amounts in induced pluripotent stem cell-derived (iPSC-derived) macrophages, and higher levels in fibroblasts and iPSC-derived neuronal precursor cells. Finally, SRSF6 binds to the PE, facilitating its inclusion. Moreover, SRSF6 knockdown or CLK inhibition restores WT NEMO expression and function in mutant cells. A recurrent deep intronic splicing mutation in IKBKG underlies a purely quantitative NEMO defect in males that is most severe in leukocytes and can be rescued by the inhibition of SRSF6 or CLK.
Keywords: Genetics; Infectious disease; Molecular diagnosis; NF-kappaB.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

