A fast adaptive algorithm for computing whole-genome homology maps
- PMID: 30423094
- PMCID: PMC6129286
- DOI: 10.1093/bioinformatics/bty597
A fast adaptive algorithm for computing whole-genome homology maps
Abstract
Motivation: Whole-genome alignment is an important problem in genomics for comparing different species, mapping draft assemblies to reference genomes and identifying repeats. However, for large plant and animal genomes, this task remains compute and memory intensive. In addition, current practical methods lack any guarantee on the characteristics of output alignments, thus making them hard to tune for different application requirements.
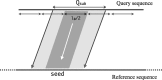
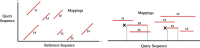
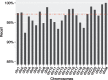
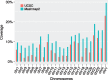
Results: We introduce an approximate algorithm for computing local alignment boundaries between long DNA sequences. Given a minimum alignment length and an identity threshold, our algorithm computes the desired alignment boundaries and identity estimates using kmer-based statistics, and maintains sufficient probabilistic guarantees on the output sensitivity. Further, to prioritize higher scoring alignment intervals, we develop a plane-sweep based filtering technique which is theoretically optimal and practically efficient. Implementation of these ideas resulted in a fast and accurate assembly-to-genome and genome-to-genome mapper. As a result, we were able to map an error-corrected whole-genome NA12878 human assembly to the hg38 human reference genome in about 1 min total execution time and <4 GB memory using eight CPU threads, achieving significant improvement in memory-usage over competing methods. Recall accuracy of computed alignment boundaries was consistently found to be >97% on multiple datasets. Finally, we performed a sensitive self-alignment of the human genome to compute all duplications of length ≥1 Kbp and ≥90% identity. The reported output achieves good recall and covers twice the number of bases than the current UCSC browser's segmental duplication annotation.
Availability and implementation: https://github.com/marbl/MashMap.
Figures



References
-
- Bailey J.A., et al. (2002) Recent segmental duplications in the human genome. Science, 297, 1003–1007. - PubMed
-
- Berman P., et al. (1999) Winnowing sequences from a database search. In: Proceedings of the Third Annual International Conference on Computational Molecular Biology. ACM, pp. 50–58. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

