Structure and Physiological Regulation of AMPK
- PMID: 30423971
- PMCID: PMC6274893
- DOI: 10.3390/ijms19113534
Structure and Physiological Regulation of AMPK
Abstract
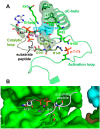
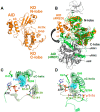
Adenosine monophosphate (AMP)-activated protein kinase (AMPK) is a heterotrimeric αβγ complex that functions as a central regulator of energy homeostasis. Energy stress manifests as a drop in the ratio of adenosine triphosphate (ATP) to AMP/ADP, which activates AMPK's kinase activity, allowing it to upregulate ATP-generating catabolic pathways and to reduce energy-consuming catabolic pathways and cellular programs. AMPK senses the cellular energy state by competitive binding of the three adenine nucleotides AMP, ADP, and ATP to three sites in its γ subunit, each, which in turn modulates the activity of AMPK's kinase domain in its α subunit. Our current understanding of adenine nucleotide binding and the mechanisms by which differential adenine nucleotide occupancies activate or inhibit AMPK activity has been largely informed by crystal structures of AMPK in different activity states. Here we provide an overview of AMPK structures, and how these structures, in combination with biochemical, biophysical, and mutational analyses provide insights into the mechanisms of adenine nucleotide binding and AMPK activity modulation.
Keywords: AID; AMPK; CBS; CaMKK2; LKB1; activation loop; energy metabolism; α-linker; αRIM; β-linker.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

