Whole-genome epidemiology, characterisation, and phylogenetic reconstruction of Staphylococcus aureus strains in a paediatric hospital
- PMID: 30424799
- PMCID: PMC6234625
- DOI: 10.1186/s13073-018-0593-7
Whole-genome epidemiology, characterisation, and phylogenetic reconstruction of Staphylococcus aureus strains in a paediatric hospital
Abstract
Background: Staphylococcus aureus is an opportunistic pathogen and a leading cause of nosocomial infections. It can acquire resistance to all the antibiotics that entered the clinics to date, and the World Health Organization defined it as a high-priority pathogen for research and development of new antibiotics. A deeper understanding of the genetic variability of S. aureus in clinical settings would lead to a better comprehension of its pathogenic potential and improved strategies to contrast its virulence and resistance. However, the number of comprehensive studies addressing clinical cohorts of S. aureus infections by simultaneously looking at the epidemiology, phylogenetic reconstruction, genomic characterisation, and transmission pathways of infective clones is currently low, thus limiting global surveillance and epidemiological monitoring.
Methods: We applied whole-genome shotgun sequencing (WGS) to 184 S. aureus isolates from 135 patients treated in different operative units of an Italian paediatric hospital over a timespan of 3 years, including both methicillin-resistant S. aureus (MRSA) and methicillin-sensitive S. aureus (MSSA) from different infection types. We typed known and unknown clones from their genomes by multilocus sequence typing (MLST), Staphylococcal Cassette Chromosome mec (SCCmec), Staphylococcal protein A gene (spa), and Panton-Valentine Leukocidin (PVL), and we inferred their whole-genome phylogeny. We explored the prevalence of virulence and antibiotic resistance genes in our cohort, and the conservation of genes encoding vaccine candidates. We also performed a timed phylogenetic investigation for a potential outbreak of a newly emerging nosocomial clone.
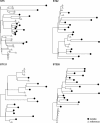
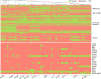
Results: The phylogeny of the 135 single-patient S. aureus isolates showed a high level of diversity, including 80 different lineages, and co-presence of local, global, livestock-associated, and hypervirulent clones. Five of these clones do not have representative genomes in public databases. Variability in the epidemiology is mirrored by variability in the SCCmec cassettes, with some novel variants of the type IV cassette carrying extra antibiotic resistances. Virulence and resistance genes were unevenly distributed across different clones and infection types, with highly resistant and lowly virulent clones showing strong association with chronic diseases, and highly virulent strains only reported in acute infections. Antigens included in vaccine formulations undergoing clinical trials were conserved at different levels in our cohort, with only a few highly prevalent genes fully conserved, potentially explaining the difficulty of developing a vaccine against S. aureus. We also found a recently diverged ST1-SCCmecIV-t127 PVL- clone suspected to be hospital-specific, but time-resolved integrative phylogenetic analysis refuted this hypothesis and suggested that this quickly emerging lineage was acquired independently by patients.
Conclusions: Whole genome sequencing allowed us to study the epidemiology and genomic repertoire of S. aureus in a clinical setting and provided evidence of its often underestimated complexity. Some virulence factors and clones are specific of disease types, but the variability and dispensability of many antigens considered for vaccine development together with the quickly changing epidemiology of S. aureus makes it very challenging to develop full-coverage therapies and vaccines. Expanding WGS-based surveillance of S. aureus to many more hospitals would allow the identification of specific strains representing the main burden of infection and therefore reassessing the efforts for the discovery of new treatments and clinical practices.
Keywords: Bacterial pathogens; Microbial epidemiology; Microbial genomics; Staphylococcus aureus.
Conflict of interest statement
Ethics approval and consent to participate
The study received ethical approval from the Paediatric Ethics Committee, Autonomous Section of the Regional Ethics Committee for Clinical Trials at the Children’s Meyer Hospital, Florence on July 1, 2014. Data collection has been performed in accordance with the Declaration of Helsinki. All patients gave written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

