Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces
- PMID: 30424821
- PMCID: PMC6234677
- DOI: 10.1186/s40168-018-0585-2
Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces
Erratum in
-
Correction to: Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces.Microbiome. 2018 Dec 4;6(1):214. doi: 10.1186/s40168-018-0609-y. Microbiome. 2018. PMID: 30514368 Free PMC article.
Abstract
Background: The International Space Station (ISS) is an ideal test bed for studying the effects of microbial persistence and succession on a closed system during long space flight. Culture-based analyses, targeted gene-based amplicon sequencing (bacteriome, mycobiome, and resistome), and shotgun metagenomics approaches have previously been performed on ISS environmental sample sets using whole genome amplification (WGA). However, this is the first study reporting on the metagenomes sampled from ISS environmental surfaces without the use of WGA. Metagenome sequences generated from eight defined ISS environmental locations in three consecutive flights were analyzed to assess the succession and persistence of microbial communities, their antimicrobial resistance (AMR) profiles, and virulence properties. Metagenomic sequences were produced from the samples treated with propidium monoazide (PMA) to measure intact microorganisms.
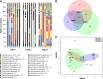
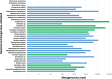
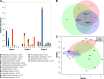
Results: The intact microbial communities detected in Flight 1 and Flight 2 samples were significantly more similar to each other than to Flight 3 samples. Among 318 microbial species detected, 46 species constituting 18 genera were common in all flight samples. Risk group or biosafety level 2 microorganisms that persisted among all three flights were Acinetobacter baumannii, Haemophilus influenzae, Klebsiella pneumoniae, Salmonella enterica, Shigella sonnei, Staphylococcus aureus, Yersinia frederiksenii, and Aspergillus lentulus. Even though Rhodotorula and Pantoea dominated the ISS microbiome, Pantoea exhibited succession and persistence. K. pneumoniae persisted in one location (US Node 1) of all three flights and might have spread to six out of the eight locations sampled on Flight 3. The AMR signatures associated with β-lactam, cationic antimicrobial peptide, and vancomycin were detected. Prominent virulence factors were cobalt-zinc-cadmium resistance and multidrug-resistance efflux pumps.
Conclusions: There was an increase in AMR and virulence gene factors detected over the period sampled, and metagenome sequences of human pathogens persisted over time. Comparative analysis of the microbial compositions of ISS with Earth analogs revealed that the ISS environmental surfaces were different in microbial composition. Metagenomics coupled with PMA treatment would help future space missions to estimate problematic risk group microbial pathogens. Cataloging AMR/virulence characteristics, succession, accumulation, and persistence of microorganisms would facilitate the development of suitable countermeasures to reduce their presence in the closed built environment.
Keywords: Built environment; Functional metagenomics; International Space Station; Metagenome; Propidium monoazide.
Conflict of interest statement
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
-
- Lax S., Smith D. P., Hampton-Marcell J., Owens S. M., Handley K. M., Scott N. M., Gibbons S. M., Larsen P., Shogan B. D., Weiss S., Metcalf J. L., Ursell L. K., Vazquez-Baeza Y., Van Treuren W., Hasan N. A., Gibson M. K., Colwell R., Dantas G., Knight R., Gilbert J. A. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345(6200):1048–1052. doi: 10.1126/science.1254529. - DOI - PMC - PubMed
-
- NRC: Committee for the Decadal Survey on Biological Physical Sciences in Space. Recapturing a future for space exploration: life and physical sciences research for a new era: The National Academies Press; 2011.
-
- Pierson D, Botkin D, Bruce R, Castro V, Smith M, Oubre C, Ott C. Microbial monitoring of the International Space Station. In: Moldenhauer J, editor. Environmental monitoring: a comprehensive handbook. River Grove: DHI Publishing, LLC; 2012.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

