The gold-standard genome of Aspergillus niger NRRL 3 enables a detailed view of the diversity of sugar catabolism in fungi
- PMID: 30425417
- PMCID: PMC6231085
- DOI: 10.1016/j.simyco.2018.10.001
The gold-standard genome of Aspergillus niger NRRL 3 enables a detailed view of the diversity of sugar catabolism in fungi
Abstract
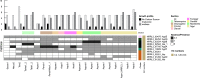
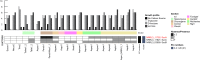
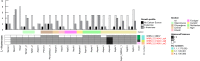
The fungal kingdom is too large to be discovered exclusively by classical genetics. The access to omics data opens a new opportunity to study the diversity within the fungal kingdom and how adaptation to new environments shapes fungal metabolism. Genomes are the foundation of modern science but their quality is crucial when analysing omics data. In this study, we demonstrate how one gold-standard genome can improve functional prediction across closely related species to be able to identify key enzymes, reactions and pathways with the focus on primary carbon metabolism. Based on this approach we identified alternative genes encoding various steps of the different sugar catabolic pathways, and as such provided leads for functional studies into this topic. We also revealed significant diversity with respect to genome content, although this did not always correlate to the ability of the species to use the corresponding sugar as a carbon source.
Keywords: Aspergillus; Genomic diversity; Gold standard genome; Sugar catabolism.
Figures









References
-
- Aguilar-Pontes M.V., de Vries R.P., Zhou M. Post-)genomics approaches in fungal research. Briefings in Functional Genomics. 2014;13:424–439. - PubMed
-
- Altschul S.F., Gish W., Miller W. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. - PubMed
-
- Benocci T., Aguilar-Pontes M.V., Kun R.S. ARA1 regulates not only L-arabinose but also D-galactose catabolism in Trichoderma reesei. FEBS Letters. 2018;592:60–70. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
