Whole Genome Sequencing for Determining the Source of Mycobacterium bovis Infections in Livestock Herds and Wildlife in New Zealand
- PMID: 30425997
- PMCID: PMC6218598
- DOI: 10.3389/fvets.2018.00272
Whole Genome Sequencing for Determining the Source of Mycobacterium bovis Infections in Livestock Herds and Wildlife in New Zealand
Abstract
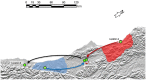
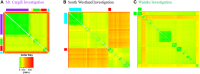
The ability to DNA fingerprint Mycobacterium bovis isolates helped to define the role of wildlife in the persistence of bovine tuberculosis in New Zealand. DNA fingerprinting results currently help to guide wildlife control measures and also aid in tracing the source of infections that result from movement of livestock. During the last 5 years we have developed the ability to distinguish New Zealand (NZ) M. bovis isolates by comparing the sequences of whole genome sequenced (WGS) M. bovis samples. WGS provides much higher resolution than our other established typing methods and greatly improves the definition of the regional localization of NZ M. bovis types. Three outbreak investigations are described and results demonstrate how WGS analysis has led to the confirmation of epidemiological sourcing of infection, to better definition of new sources of infection by ruling out other possible sources, and has revealed probable wildlife infection in an area considered to be free of infected wildlife. The routine use of WGS analyses for sourcing new M. bovis infections will be an important component of the strategy employed to eradicate bovine TB from NZ livestock and wildlife.
Keywords: Mycobacterium bovis; New Zealand; bovine tuberculosis control; epidemiology; molecular fingerprint; whole genome sequencing.
Figures






Similar articles
-
Using whole genome sequencing to investigate transmission in a multi-host system: bovine tuberculosis in New Zealand.BMC Genomics. 2017 Feb 16;18(1):180. doi: 10.1186/s12864-017-3569-x. BMC Genomics. 2017. PMID: 28209138 Free PMC article.
-
Disease management at the wildlife-livestock interface: Using whole-genome sequencing to study the role of elk in Mycobacterium bovis transmission in Michigan, USA.Mol Ecol. 2019 May;28(9):2192-2205. doi: 10.1111/mec.15061. Epub 2019 May 10. Mol Ecol. 2019. PMID: 30807679
-
Use of whole-genome sequencing and evaluation of the apparent sensitivity and specificity of antemortem tuberculosis tests in the investigation of an unusual outbreak of Mycobacterium bovis infection in a Michigan dairy herd.J Am Vet Med Assoc. 2017 Jul 15;251(2):206-216. doi: 10.2460/javma.251.2.206. J Am Vet Med Assoc. 2017. PMID: 28671497
-
Feral ferrets (Mustela furo) as hosts and sentinels of tuberculosis in New Zealand.N Z Vet J. 2015 Jun;63 Suppl 1(sup1):42-53. doi: 10.1080/00480169.2014.981314. Epub 2015 Mar 10. N Z Vet J. 2015. PMID: 25495945 Free PMC article. Review.
-
Mycobacterium bovis: From Genotyping to Genome Sequencing.Microorganisms. 2020 May 3;8(5):667. doi: 10.3390/microorganisms8050667. Microorganisms. 2020. PMID: 32375210 Free PMC article. Review.
Cited by
-
Genomic and temporal analyses of Mycobacterium bovis in southern Brazil.Microb Genom. 2021 May;7(5):000569. doi: 10.1099/mgen.0.000569. Microb Genom. 2021. PMID: 34016251 Free PMC article.
-
Generalized tuberculosis due to Mycobacterium caprae in a red fox phylogenetically related to livestock breakdowns.BMC Vet Res. 2022 Sep 20;18(1):352. doi: 10.1186/s12917-022-03454-7. BMC Vet Res. 2022. PMID: 36127697 Free PMC article.
-
Performance and Agreement Between WGS Variant Calling Pipelines Used for Bovine Tuberculosis Control: Toward International Standardization.Front Vet Sci. 2021 Dec 14;8:780018. doi: 10.3389/fvets.2021.780018. eCollection 2021. Front Vet Sci. 2021. PMID: 34970617 Free PMC article.
-
Whole-Genome SNP Analysis Identifies Putative Mycobacterium bovis Transmission Clusters in Livestock and Wildlife in Catalonia, Spain.Microorganisms. 2021 Jul 30;9(8):1629. doi: 10.3390/microorganisms9081629. Microorganisms. 2021. PMID: 34442709 Free PMC article.
-
Enhanced analysis of the genomic diversity of Mycobacterium bovis in Great Britain to aid control of bovine tuberculosis.Front Microbiol. 2025 Mar 25;16:1515906. doi: 10.3389/fmicb.2025.1515906. eCollection 2025. Front Microbiol. 2025. PMID: 40201440 Free PMC article.
References
-
- Livingstone PG, Hancox N, Nugent G, de Lisle GW. Toward eradication: the effect of Mycobacterium bovis infection in wildlife on the evolution and future direction of bovine tuberculosis management in New Zealand. N Z Vet J. (2015) 63 (Suppl. 1):4–18. 10.1080/00480169.2014.971082 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

