Long-term live imaging of the Drosophila adult midgut reveals real-time dynamics of division, differentiation and loss
- PMID: 30427308
- PMCID: PMC6277200
- DOI: 10.7554/eLife.36248
Long-term live imaging of the Drosophila adult midgut reveals real-time dynamics of division, differentiation and loss
Abstract
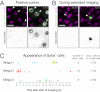
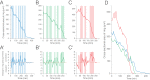
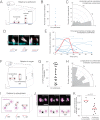
Organ renewal is governed by the dynamics of cell division, differentiation and loss. To study these dynamics in real time, we present a platform for extended live imaging of the adult Drosophila midgut, a premier genetic model for stem-cell-based organs. A window cut into a living animal allows the midgut to be imaged while intact and physiologically functioning. This approach prolongs imaging sessions to 12-16 hr and yields movies that document cell and tissue dynamics at vivid spatiotemporal resolution. By applying a pipeline for movie processing and analysis, we uncover new and intriguing cell behaviors: that mitotic stem cells dynamically re-orient, that daughter cells use slow kinetics of Notch activation to reach a fate-specifying threshold, and that enterocytes extrude via ratcheted constriction of a junctional ring. By enabling real-time study of midgut phenomena that were previously inaccessible, our platform opens a new realm for dynamic understanding of adult organ renewal.
Keywords: D. melanogaster; adult tissue homeostasis; apical extrusion; differentiation; epithelium; imaging; regenerative medicine; stem cells.
© 2018, Martin et al.
Conflict of interest statement
JM, ES, PM, LJ, SB, XD, LO No competing interests declared
Figures







References
-
- Arganda-Carreras I, Sorzano COS, Marabini R, Carazo JM, Ortiz-de-Solorzano C, Kybic J. Consistent and elastic registration of histological sections using vector-spline regularization. In: Beichel RR, Sonka M, editors. Computer Vision Approaches to Medical Image Analysis. CVAMIA 2006. Lecture Notes in Computer Science. 4241. Berlin, Heidelberg: Springer; 2006a. pp. 85–95.
-
- Arganda-Carreras I, Sorzano COS, Marabini R, Carazo JM, Ortiz-de-Solorzano C, Kybic J. Consistent and Elastic Registration of Histological Sections Using Vector-Spline Regularization. Lecture Notes in Computer Science. 2006b:85–95.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

