Functional and anatomical relationships between the medial precentral cortex, dorsal striatum, and head direction cell circuitry. II. Neuroanatomical studies
- PMID: 30427743
- PMCID: PMC6397393
- DOI: 10.1152/jn.00144.2018
Functional and anatomical relationships between the medial precentral cortex, dorsal striatum, and head direction cell circuitry. II. Neuroanatomical studies
Abstract
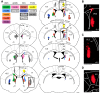
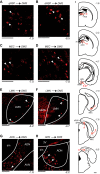
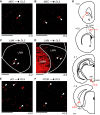
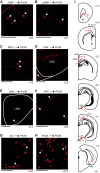
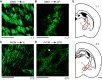
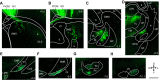
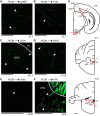
An animal's directional heading within its environment is encoded by the activity of head direction (HD) cells. In rodents, these neurons are found primarily within the limbic system in the interconnected structures that form the limbic HD circuit. In our accompanying report in this issue, we describe two HD cell populations located outside of this circuit in the medial precentral cortex (PrCM) and dorsal striatum (DS). These extralimbic areas receive their HD signals from the limbic system but do not provide critical input or feedback to limbic HD cells (Mehlman ML, Winter SS, Valerio S, Taube JS. J Neurophysiol 121: 350-370, 2019.). In this report, we complement our previous lesion and recording experiments with a series of neuroanatomical tracing studies in rats designed to examine patterns of connectivity between the PrCM, DS, limbic HD circuit, and related spatial processing circuitry. Retrograde tracing revealed that the DS receives direct input from numerous structures known to contain HD cells and/or other spatially tuned cell types. Importantly, these projections preferentially target and converge within the most medial portion of the DS, the same area in which we previously recorded HD cells. The PrCM receives direct input from a subset of these spatial processing structures. Anterograde tracing identified indirect pathways that could permit the PrCM and DS to convey self-motion information to the limbic HD circuit. These tracing studies reveal the anatomical basis for the functional relationships observed in our lesion and recording experiments. Collectively, these findings expand our understanding of how spatial processing circuitry functionally and anatomically extends beyond the limbic system into the PrCM and DS. NEW & NOTEWORTHY Head direction (HD) cells are located primarily within the limbic system, but small populations of extralimbic HD cells are found in the medial precentral cortex (PrCM) and dorsal striatum (DS). The neuroanatomical tracing experiments reported here explored the pathways capable of transmitting the HD signal to these extralimbic areas. We found that projections arising from numerous spatial processing structures converge within portions of the PrCM and DS that contain HD cells.
Keywords: dorsal striatum; head direction cell; limbic system; medial precentral cortex; navigation; spatial cognition.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

