Characterization of Haartman Institute snake virus-1 (HISV-1) and HISV-like viruses-The representatives of genus Hartmanivirus, family Arenaviridae
- PMID: 30427944
- PMCID: PMC6261641
- DOI: 10.1371/journal.ppat.1007415
Characterization of Haartman Institute snake virus-1 (HISV-1) and HISV-like viruses-The representatives of genus Hartmanivirus, family Arenaviridae
Abstract
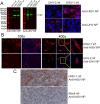
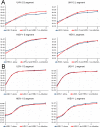
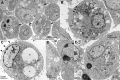
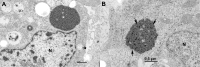
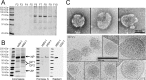
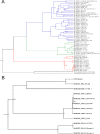
The family Arenaviridae comprises three genera, Mammarenavirus, Reptarenavirus and the most recently added Hartmanivirus. Arenaviruses have a bisegmented genome with ambisense coding strategy. For mammarenaviruses and reptarenaviruses the L segment encodes the Z protein (ZP) and the RNA-dependent RNA polymerase, and the S segment encodes the glycoprotein precursor and the nucleoprotein. Herein we report the full length genome and characterization of Haartman Institute snake virus-1 (HISV-1), the putative type species of hartmaniviruses. The L segment of HISV-1 lacks an open-reading frame for ZP, and our analysis of purified HISV-1 particles by SDS-PAGE and electron microscopy further support the lack of ZP. Since we originally identified HISV-1 in co-infection with a reptarenavirus, one could hypothesize that co-infecting reptarenavirus provides the ZP to complement HISV-1. However, we observed that co-infection does not markedly affect the amount of hartmanivirus or reptarenavirus RNA released from infected cells in vitro, indicating that HISV-1 does not benefit from reptarenavirus ZP. Furthermore, we succeeded in generating a pure HISV-1 isolate showing the virus to replicate without ZP. Immunofluorescence and ultrastructural studies demonstrate that, unlike reptarenaviruses, HISV-1 does not produce the intracellular inclusion bodies typical for the reptarenavirus-induced boid inclusion body disease (BIBD). While we observed HISV-1 to be slightly cytopathic for cultured boid cells, the histological and immunohistological investigation of HISV-positive snakes showed no evidence of a pathological effect. The histological analyses also revealed that hartmaniviruses, unlike reptarenaviruses, have a limited tissue tropism. By nucleic acid sequencing, de novo genome assembly, and phylogenetic analyses we identified additional four hartmanivirus species. Finally, we screened 71 individuals from a collection of snakes with BIBD by RT-PCR and found 44 to carry hartmaniviruses. These findings suggest that harmaniviruses are common in captive snake populations, but their relevance and pathogenic potential needs yet to be revealed.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel RN, Clawson AN, et al. (2015) Past, present, and future of arenavirus taxonomy Arch Virol 160(7): 1851–1874. 10.1007/s00705-015-2418-y - DOI - PubMed
-
- Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, et al. (2012) Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: Candidate etiological agents for snake inclusion body disease. MBio 3(4): e00180–12. 10.1128/mBio.00180-12 - DOI - PMC - PubMed
-
- Bodewes R, Kik MJ, Raj VS, Schapendonk CM, Haagmans BL, et al. (2013) Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in the netherlands. J Gen Virol 94(Pt 6): 1206–1210. 10.1099/vir.0.051995-0 - DOI - PubMed
-
- Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, et al. (2013) Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J Virol 87(20): 10918–10935. 10.1128/JVI.01123-13 - DOI - PMC - PubMed
-
- Bodewes R, Raj VS, Kik MJ, Schapendonk CM, Haagmans BL, et al. (2014) Updated phylogenetic analysis of arenaviruses detected in boid snakes. J Virol 88(2): 1399–1400. 10.1128/JVI.02753-13 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

