A Drug-Side Effect Context-Sensitive Network approach for drug target prediction
- PMID: 30428013
- PMCID: PMC6581434
- DOI: 10.1093/bioinformatics/bty906
A Drug-Side Effect Context-Sensitive Network approach for drug target prediction
Abstract
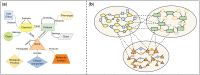
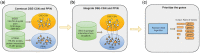
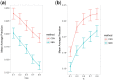
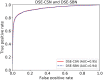
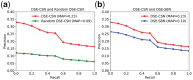
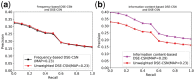
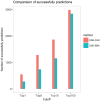
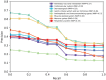
Summary: Computational drug target prediction has become an important process in drug discovery. Network-based approaches are commonly used in computational drug-target interaction (DTI) prediction. Existing network-based approaches are limited in capturing the contextual information on how diseases, drugs and genes are connected. Here, we proposed a context-sensitive network (CSN) model for DTI prediction by modeling contextual drug phenotypic relationships. We constructed a Drug-Side Effect Context-Sensitive Network (DSE-CSN) of 139 760 drug-side effect pairs, representing 1480 drugs and 5868 side effects. We also built a protein-protein interaction network (PPIN) of 15 267 gene nodes and 178 972 weighted edges. A heterogeneous network was built by connecting the DSE-CSN and the PPIN through 3684 known DTIs. For each drug on the DSE-CSN, its genetic targets were predicted and prioritized using a network-based ranking algorithm. Our approach was evaluated in both de novo and leave-one-out cross-validation analysis using known DTIs as the gold standard. We compared our DSE-CSN-based model to the traditional similarity-based network (SBN)-based prediction model. The results suggested that the DSE-CSN-based model was able to rank known DTIs highly. In a de novo cross-validation, the area under the receiver operating characteristic (ROC) curve was 0.95. In a leave-one-out cross-validation, the average rank was top 3.2% for known DTIs. When it was compared to the SBN-based model using the Precision-Recall curve, our CSN-based model achieved a higher mean average precision (MAP) (0.23 versus 0.19, P-value<1e-4) in a de novo cross-validation analysis. We further improved the CSN-based DTI prediction by differentially weighting the drug-side effect pairs on the network and showed a significant improvement of the MAP (0.29 versus 0.23, P-value<1e-4). We also showed that the CSN-based model consistently achieved better performances than the traditional SBN-based model across different drug classes. Moreover, we demonstrated that our novel DTI predictions can be supported by published literature. In summary, the CSN-based model, by modeling the context-specific inter-relationships among drugs and side effects, has a high potential in drug target prediction.
Availability and implementation: nlp/case/edu/public/data/DSE/CSN_DTI.
© The Author(s) 2018. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

Similar articles
-
Context-sensitive network-based disease genetics prediction and its implications in drug discovery.Bioinformatics. 2017 Apr 1;33(7):1031-1039. doi: 10.1093/bioinformatics/btw737. Bioinformatics. 2017. PMID: 28062449 Free PMC article.
-
Combining phenome-driven drug-target interaction prediction with patients' electronic health records-based clinical corroboration toward drug discovery.Bioinformatics. 2020 Jul 1;36(Suppl_1):i436-i444. doi: 10.1093/bioinformatics/btaa451. Bioinformatics. 2020. PMID: 32657406 Free PMC article.
-
MultiDTI: drug-target interaction prediction based on multi-modal representation learning to bridge the gap between new chemical entities and known heterogeneous network.Bioinformatics. 2021 Dec 7;37(23):4485-4492. doi: 10.1093/bioinformatics/btab473. Bioinformatics. 2021. PMID: 34180970
-
Comparative analysis of network-based approaches and machine learning algorithms for predicting drug-target interactions.Methods. 2022 Feb;198:19-31. doi: 10.1016/j.ymeth.2021.10.007. Epub 2021 Nov 1. Methods. 2022. PMID: 34737033 Review.
-
Open-source chemogenomic data-driven algorithms for predicting drug-target interactions.Brief Bioinform. 2019 Jul 19;20(4):1465-1474. doi: 10.1093/bib/bby010. Brief Bioinform. 2019. PMID: 29420684 Free PMC article. Review.
Cited by
-
Identifying the serious clinical outcomes of adverse reactions to drugs by a multi-task deep learning framework.Commun Biol. 2023 Aug 24;6(1):870. doi: 10.1038/s42003-023-05243-w. Commun Biol. 2023. PMID: 37620651 Free PMC article.
-
Drug repurposing for opioid use disorders: integration of computational prediction, clinical corroboration, and mechanism of action analyses.Mol Psychiatry. 2021 Sep;26(9):5286-5296. doi: 10.1038/s41380-020-01011-y. Epub 2021 Jan 11. Mol Psychiatry. 2021. PMID: 33432189 Free PMC article.
-
NeuRank: learning to rank with neural networks for drug-target interaction prediction.BMC Bioinformatics. 2021 Nov 26;22(1):567. doi: 10.1186/s12859-021-04476-y. BMC Bioinformatics. 2021. PMID: 34836495 Free PMC article.
-
Supervised Analysis for Phenotype Identification: The Case of Heart Failure Ejection Fraction Class.Bioengineering (Basel). 2021 Jun 21;8(6):85. doi: 10.3390/bioengineering8060085. Bioengineering (Basel). 2021. PMID: 34205745 Free PMC article.
-
Quantitative prediction model for affinity of drug-target interactions based on molecular vibrations and overall system of ligand-receptor.BMC Bioinformatics. 2021 Oct 14;22(1):497. doi: 10.1186/s12859-021-04389-w. BMC Bioinformatics. 2021. PMID: 34649499 Free PMC article.
References
-
- Campillos M. et al. (2008) Drug target identification using side-effect similarity. Science, 321, 263–266. - PubMed
-
- Chen X. et al. (2012) Drug–target interaction prediction by random walk on the heterogeneous network. Mol. BioSyst., 8, 1970–1978. - PubMed
-
- Chen X. et al. (2016) Drug–target interaction prediction: databases, web servers and computational models. Brief. Bioinf., 17, 696–712. - PubMed

