Methods for Estimating Demography and Detecting Between-Locus Differences in the Effective Population Size and Mutation Rate
- PMID: 30428070
- PMCID: PMC6409433
- DOI: 10.1093/molbev/msy212
Methods for Estimating Demography and Detecting Between-Locus Differences in the Effective Population Size and Mutation Rate
Abstract
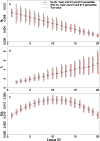
It is known that the effective population size (Ne) and the mutation rate (u) vary across the genome. Here, we show that ignoring this heterogeneity may lead to biased estimates of past demography. To solve the problem, we develop new methods for jointly inferring past changes in population size and detecting variation in Ne and u between loci. These methods rely on either polymorphism data alone or both polymorphism and divergence data. In addition to inferring demography, we can use the methods to study a variety of questions: 1) comparing sex chromosomes with autosomes (for finding evidence for male-driven evolution, an unequal sex ratio, or sex-biased demographic changes) and 2) analyzing multilocus data from within autosomes or sex chromosomes (for studying determinants of variability in Ne and u). Simulations suggest that the methods can provide accurate parameter estimates and have substantial statistical power for detecting difference in Ne and u. As an example, we use the methods to analyze a polymorphism data set from Drosophila simulans. We find clear evidence for rapid population expansion. The results also indicate that the autosomes have a higher mutation rate than the X chromosome and that the sex ratio is probably female-biased. The new methods have been implemented in a user-friendly package.
Figures
References
-
- Allendorf FW, Hohenlohe PA, Luikart G.. 2010. Genomics and the future of conservation genetics. Nat Rev Genet. 1110: 697–709. - PubMed
-
- Bachtrog D. 2008. Evidence for male-driven evolution in Drosophila. Mol Biol Evol. 254: 617–619. - PubMed
-
- Bank C, Ewing GB, Ferrer-Admettla A, Foll M, Jensen JD.. 2014. Thinking too positive? Revisiting current methods of population genetic selection inference. Trends Genet. 3012: 540–546. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

