Autophagy Ablation in Adipocytes Induces Insulin Resistance and Reveals Roles for Lipid Peroxide and Nrf2 Signaling in Adipose-Liver Crosstalk
- PMID: 30428342
- PMCID: PMC6802939
- DOI: 10.1016/j.celrep.2018.10.040
Autophagy Ablation in Adipocytes Induces Insulin Resistance and Reveals Roles for Lipid Peroxide and Nrf2 Signaling in Adipose-Liver Crosstalk
Abstract
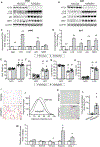
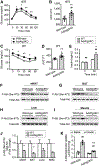
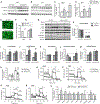
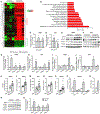
Autophagy is a homeostatic cellular process involved in the degradation of long-lived or damaged cellular components. The role of autophagy in adipogenesis is well recognized, but its role in mature adipocyte function is largely unknown. We show that the autophagy proteins Atg3 and Atg16L1 are required for proper mitochondrial function in mature adipocytes. In contrast to previous studies, we found that post-developmental ablation of autophagy causes peripheral insulin resistance independently of diet or adiposity. Finally, lack of adipocyte autophagy reveals cross talk between fat and liver, mediated by lipid peroxide-induced Nrf2 signaling. Our data reveal a role for autophagy in preventing lipid peroxide formation and its transfer in insulin-sensitive peripheral tissues.
Keywords: adipocytes; adiponectin; adipose tissue; autophagy; inflammation; insulin resistance; lipid peroxide; mitochondria.
Copyright © 2018 University of Utah. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures
References
-
- Boudina S, and Graham TE (2014). Mitochondrial function/dysfunction in white adipose tissue. Exp. Physiol 99, 1168–1178. - PubMed
-
- Demozay D, Mas JC, Rocchi S, and Van Obberghen E (2008). FALDH reverses the deleterious action of oxidative stress induced by lipid peroxidation product 4-hydroxynonenal on insulin signaling in 3T3-L1 adipocytes. Diabetes 57, 1216–1226. - PubMed
-
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, and Talalay P (2002). Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 99, 11908–11913. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

