TP63-Mediated Enhancer Reprogramming Drives the Squamous Subtype of Pancreatic Ductal Adenocarcinoma
- PMID: 30428345
- PMCID: PMC6296757
- DOI: 10.1016/j.celrep.2018.10.051
TP63-Mediated Enhancer Reprogramming Drives the Squamous Subtype of Pancreatic Ductal Adenocarcinoma
Abstract
The aberrant expression of squamous lineage markers in pancreatic ductal adenocarcinoma (PDA) has been correlated with poor clinical outcomes. However, the functional role of this putative transdifferentiation event in PDA pathogenesis remains unclear. Here, we show that expression of the transcription factor TP63 (ΔNp63) is sufficient to install and sustain the enhancer landscape and transcriptional signature of the squamous lineage in human PDA cells. We also demonstrate that TP63-driven enhancer reprogramming promotes aggressive tumor phenotypes, including enhanced cell motility and invasion, and an accelerated growth of primary PDA tumors and metastases in vivo. This process ultimately leads to a powerful addiction of squamous PDA cells to continuous TP63 expression. Our study demonstrates the functional significance of squamous transdifferentiation in PDA and reveals TP63-based reprogramming as an experimental tool for investigating mechanisms and vulnerabilities linked to this aberrant cell fate transition.
Keywords: H3K27ac; TP63; chromatin; enhancer; enhancer reprogramming; master regulator; squamous.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
C.R.V. is an advisor to KSQ Therapeutics and receives research funding from Boehringer Ingelheim. D.A.T. is a member of the scientific advisory boards of Surface Oncology, Leap Therapeutics, MabImmune, VividBiosciences, and Bethyl Antibodies. D.A.T. has stock ownership in Surface Oncology and Leap Therapeutics. D.A.T. receives research support from ONO and Fibrogen.
Figures




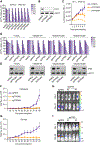
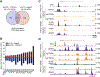
References
-
- Aiello NM, Rhim AD, and Stanger BZ (2016). Orthotopic injection of pancreatic cancer cells. Cold Spring Harb. Protoc 2016, pdb.prot078360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

