Broad-Spectrum Antiviral Activity of an Ankyrin Repeat Protein on Viral Assembly against Chimeric NL4-3 Viruses Carrying Gag/PR Derived from Circulating Strains among Northern Thai Patients
- PMID: 30428529
- PMCID: PMC6265948
- DOI: 10.3390/v10110625
Broad-Spectrum Antiviral Activity of an Ankyrin Repeat Protein on Viral Assembly against Chimeric NL4-3 Viruses Carrying Gag/PR Derived from Circulating Strains among Northern Thai Patients
Abstract
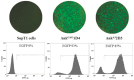
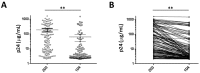
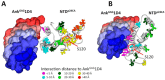
Certain proteins have demonstrated proficient human immunodeficiency virus (HIV-1) life cycle disturbance. Recently, the ankyrin repeat protein targeting the HIV-1 capsid, AnkGAG1D4, showed a negative effect on the viral assembly of the HIV-1NL4-3 laboratory strain. To extend its potential for future clinical application, the activity of AnkGAG1D4 in the inhibition of other HIV-1 circulating strains was evaluated. Chimeric NL4-3 viruses carrying patient-derived Gag/PR-coding regions were generated from 131 antiretroviral drug-naïve HIV-1 infected individuals in northern Thailand during 2001⁻2012. SupT1, a stable T-cell line expressing AnkGAG1D4 and ankyrin non-binding control (AnkA32D3), were challenged with these chimeric viruses. The p24CA sequences were analysed and classified using the K-means clustering method. Among all the classes of virus classified using the p24CA sequences, SupT1/AnkGAG1D4 demonstrated significantly lower levels of p24CA than SupT1/AnkA32D3, which was found to correlate with the syncytia formation. This result suggests that AnkGAG1D4 can significantly interfere with the chimeric viruses derived from patients with different sequences of the p24CA domain. It supports the possibility of ankyrin-based therapy as a broad alternative therapeutic molecule for HIV-1 gene therapy in the future.
Keywords: Gag polyprotein; HIV-1; ankyrin; protein therapy; virus assembly inhibitor.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production.Biomolecules. 2021 Sep 30;11(10):1437. doi: 10.3390/biom11101437. Biomolecules. 2021. PMID: 34680070 Free PMC article.
-
Antiviral activity of recombinant ankyrin targeted to the capsid domain of HIV-1 Gag polyprotein.Retrovirology. 2012 Feb 20;9:17. doi: 10.1186/1742-4690-9-17. Retrovirology. 2012. PMID: 22348230 Free PMC article.
-
Crystal structure of an antiviral ankyrin targeting the HIV-1 capsid and molecular modeling of the ankyrin-capsid complex.J Comput Aided Mol Des. 2014 Aug;28(8):869-84. doi: 10.1007/s10822-014-9772-9. Epub 2014 Jul 5. J Comput Aided Mol Des. 2014. PMID: 24997121
-
[Assembly and maturation of HIV-1--targets for new anti-HIV compounds].Mol Gen Mikrobiol Virusol. 2005;(1):18-23. Mol Gen Mikrobiol Virusol. 2005. PMID: 15790028 Review. Russian.
-
HIV type 1 Gag as a target for antiviral therapy.AIDS Res Hum Retroviruses. 2012 Jan;28(1):54-75. doi: 10.1089/AID.2011.0230. Epub 2011 Sep 21. AIDS Res Hum Retroviruses. 2012. PMID: 21848364 Free PMC article. Review.
Cited by
-
Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production.Biomolecules. 2021 Sep 30;11(10):1437. doi: 10.3390/biom11101437. Biomolecules. 2021. PMID: 34680070 Free PMC article.
-
Dimeric Ankyrin with Inverted Module Promotes Bifunctional Property in Capturing Capsid to Impede HIV-1 Replication.Int J Mol Sci. 2023 Mar 9;24(6):5266. doi: 10.3390/ijms24065266. Int J Mol Sci. 2023. PMID: 36982337 Free PMC article.
-
Specific Interaction of DARPin with HIV-1 CANTD Disturbs the Distribution of Gag, RNA Packaging, and Tetraspanin Remodelling in the Membrane.Viruses. 2022 Apr 15;14(4):824. doi: 10.3390/v14040824. Viruses. 2022. PMID: 35458554 Free PMC article.
-
Designed Ankyrin Repeat Proteins: A New Class of Viral Entry Inhibitors.Viruses. 2022 Oct 12;14(10):2242. doi: 10.3390/v14102242. Viruses. 2022. PMID: 36298797 Free PMC article. Review.
-
Occupation of a thermoresistant-scaffold (αRep) at SP1-NC cleavage site disturbs the function of HIV-1 protease.Biosci Rep. 2020 Jun 26;40(6):BSR20201131. doi: 10.1042/BSR20201131. Biosci Rep. 2020. PMID: 32519747 Free PMC article.
References
-
- Lemke C.T., Titolo S., von Schwedler U., Goudreau N., Mercier J.F., Wardrop E., Faucher A.M., Coulombe R., Banik S.S., Fader L., et al. Distinct effects of two HIV-1 capsid assembly inhibitor families that bind the same site within the N-terminal domain of the viral CA protein. J. Virol. 2012;86:6643–6655. doi: 10.1128/JVI.00493-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

