Revisiting Bacterial Ubiquitin Ligase Effectors: Weapons for Host Exploitation
- PMID: 30428531
- PMCID: PMC6274744
- DOI: 10.3390/ijms19113576
Revisiting Bacterial Ubiquitin Ligase Effectors: Weapons for Host Exploitation
Abstract
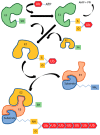
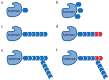
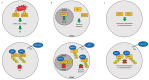
Protein ubiquitylation plays a central role in eukaryotic cell physiology. It is involved in several regulatory processes, ranging from protein folding or degradation, subcellular localization of proteins, vesicular trafficking and endocytosis to DNA repair, cell cycle, innate immunity, autophagy, and apoptosis. As such, it is reasonable that pathogens have developed a way to exploit such a crucial system to enhance their virulence against the host. Hence, bacteria have evolved a wide range of effectors capable of mimicking the main players of the eukaryotic ubiquitin system, in particular ubiquitin ligases, by interfering with host physiology. Here, we give an overview of this topic and, in particular, we detail and discuss the mechanisms developed by pathogenic bacteria to hijack the host ubiquitination system for their own benefit.
Keywords: T3SS; T4SS; ubiquitin ligase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Pisano A., Ceglia S., Palmieri C., Vecchio E., Fiume G., de Laurentiis A., Mimmi S., Falcone C., Iaccino E., Scialdone A., et al. Crl3ibtk regulates the tumor suppressor pdcd4 through ubiquitylation coupled to proteasomal degradation. J. Biol. Chem. 2015;290:13958–13971. doi: 10.1074/jbc.M114.634535. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

