Impact of polymorphisms in DNA repair genes XPD, hOGG1 and XRCC4 on colorectal cancer risk in a Chinese Han Population
- PMID: 30429230
- PMCID: PMC6331672
- DOI: 10.1042/BSR20181074
Impact of polymorphisms in DNA repair genes XPD, hOGG1 and XRCC4 on colorectal cancer risk in a Chinese Han Population
Abstract
Background: This research aimed to study the associations between XPD (G751A, rs13181), hOGG1 (C326G, rs1052133) and XRCC4 (G1394T, rs6869366) gene polymorphisms and the risk of colorectal cancer (CRC) in a Chinese Han population.
Method: A total of 225 Chinese Han patients with CRC were selected as the study group, and 200 healthy subjects were recruited as the control group. The polymorphisms of XPD G751A, hOGG1 C326G and XRCC4 G1394T loci were detected by the RFLP-PCR technique in the peripheral blood of all subjects.
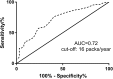
Results: Compared with individuals carrying the XPD751 GG allele, the A allele carriers (GA/AA) had a significantly increased risk of CRC (adjusted OR = 2.109, 95%CI = 1.352-3.287, P=0.003). Similarly, the G allele (CG/GG) of hOGG1 C326G locus conferred increased susceptibility to CRC (adjusted OR = 2.654, 95%CI = 1.915-3.685, P<0.001). In addition, the T allele carriers (GT/TT) of the XRCC4 G1394T locus have an increased risk of developing CRC (adjusted OR = 4.512, 95%CI = 2.785-7.402, P<0.001). The risk of CRC was significantly increased in individuals with both the XPD locus A allele and the hOGG1 locus G allele (adjusted OR = 1.543, 95%CI = 1.302-2.542, P=0.002). Furthermore, individuals with both the hOGG1 locus G allele and the XRCC4 locus T allele were predisposed to CRC development (adjusted OR = 3.854, 95%CI = 1.924-7.123, P<0.001). The risks of CRC in XPD gene A allele carriers (GA/AA) (adjusted OR = 1.570, 95%CI = 1.201-1.976, P=0.001), hOGG1 gene G allele carriers (CG/GG) (adjusted OR = 3.031, 95%CI = 2.184-4.225, P<0.001) and XRCC4 gene T allele carriers (GT/TT) (adjusted OR = 2.793, 95%CI = 2.235-3.222, P<0.001) were significantly higher in patients who smoked ≥16 packs/year.
Conclusion: Our results suggest that XPD G751A, hOGG1 C326G and XRCC4 G1394T gene polymorphisms might play an important role in colorectal carcinogenesis and increase the risk of developing CRC in the Chinese Han population. The interaction between smoking and these gene polymorphisms would increase the risk of CRC.
Keywords: Colorectal cancer; DNA repair genes; gene polymorphism.
© 2019 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
Common polymorphisms in the XPD and hOGG1 genes are not associated with the risk of colorectal cancer in a Polish population.Tohoku J Exp Med. 2009 Jul;218(3):185-91. doi: 10.1620/tjem.218.185. Tohoku J Exp Med. 2009. PMID: 19561388
-
Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population.Curr Med Res Opin. 2011 Jul;27(7):1295-302. doi: 10.1185/03007995.2011.573544. Epub 2011 May 12. Curr Med Res Opin. 2011. PMID: 21561390
-
X-ray repair cross-complementing group 4 (XRCC4) promoter -1394( *)T-related genotype, but not XRCC4 codon 247/intron 3 or xeroderma pigmentosum group D codon 312, 751/promoter -114, polymorphisms are correlated with higher susceptibility to myoma.Fertil Steril. 2008 Oct;90(4 Suppl):1417-23. doi: 10.1016/j.fertnstert.2007.09.038. Epub 2008 Jan 3. Fertil Steril. 2008. PMID: 18177646
-
Association Between the hOGG1 1245C>G (rs1052133) Polymorphism and Susceptibility to Colorectal Cancer: a Meta-analysis Based on 7010 Cases and 10,674 Controls.J Gastrointest Cancer. 2021 Jun;52(2):389-398. doi: 10.1007/s12029-020-00532-7. Epub 2020 Oct 6. J Gastrointest Cancer. 2021. PMID: 33025423 Review.
-
The association between XPD rs13181 and rs1799793 polymorphism and oral cancer risk: evidence from a meta-analysis.BMC Cancer. 2024 Jun 15;24(1):738. doi: 10.1186/s12885-024-12503-3. BMC Cancer. 2024. PMID: 38879503 Free PMC article.
Cited by
-
Nucleotide excision repair gene polymorphisms and hepatoblastoma susceptibility in Eastern Chinese children: A five-center case-control study.Chin J Cancer Res. 2024 Jun 30;36(3):298-305. doi: 10.21147/j.issn.1000-9604.2024.03.06. Chin J Cancer Res. 2024. PMID: 38988482 Free PMC article.
-
The Association of Single-Nucleotide Polymorphism rs13181 in ERCC2 with Risk and Prognosis of Nasopharyngeal Carcinoma in an Endemic Chinese Population.Pharmgenomics Pers Med. 2021 Mar 17;14:359-367. doi: 10.2147/PGPM.S296215. eCollection 2021. Pharmgenomics Pers Med. 2021. PMID: 33762840 Free PMC article.
-
Polymorphisms of nucleotide excision repair genes associated with colorectal cancer risk: Meta-analysis and trial sequential analysis.Front Genet. 2022 Oct 31;13:1009938. doi: 10.3389/fgene.2022.1009938. eCollection 2022. Front Genet. 2022. PMID: 36386844 Free PMC article.
-
The Contribution of DNA Ligase 4 Polymorphisms to Colorectal Cancer.In Vivo. 2024 Jan-Feb;38(1):127-133. doi: 10.21873/invivo.13419. In Vivo. 2024. PMID: 38148049 Free PMC article.
-
XRCC4 rs28360071 intronic variant is associated with increased risk for infant acute lymphoblastic leukemia with KMT2A rearrangements.Genet Mol Biol. 2020 Dec 2;43(4):e20200160. doi: 10.1590/1678-4685-GMB-2020-0160. eCollection 2020. Genet Mol Biol. 2020. PMID: 33270074 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

