Genetic variant at coronary artery disease and ischemic stroke locus 1p32.2 regulates endothelial responses to hemodynamics
- PMID: 30429326
- PMCID: PMC6275533
- DOI: 10.1073/pnas.1810568115
Genetic variant at coronary artery disease and ischemic stroke locus 1p32.2 regulates endothelial responses to hemodynamics
Abstract
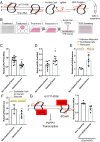
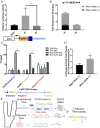
Biomechanical cues dynamically control major cellular processes, but whether genetic variants actively participate in mechanosensing mechanisms remains unexplored. Vascular homeostasis is tightly regulated by hemodynamics. Exposure to disturbed blood flow at arterial sites of branching and bifurcation causes constitutive activation of vascular endothelium contributing to atherosclerosis, the major cause of coronary artery disease (CAD) and ischemic stroke (IS). Conversely, unidirectional flow promotes quiescent endothelium. Genome-wide association studies (GWAS) have identified chromosome 1p32.2 as strongly associated with CAD/IS; however, the causal mechanism related to this locus remains unknown. Using statistical analyses, assay of transposase accessible chromatin with whole-genome sequencing (ATAC-seq), H3K27ac/H3K4me2 ChIP with whole-genome sequencing (ChIP-seq), and CRISPR interference in human aortic endothelial cells (HAECs), our results demonstrate that rs17114036, a common noncoding polymorphism at 1p32.2, is located in an endothelial enhancer dynamically regulated by hemodynamics. CRISPR-Cas9-based genome editing shows that rs17114036-containing region promotes endothelial quiescence under unidirectional shear stress by regulating phospholipid phosphatase 3 (PLPP3). Chromatin accessibility quantitative trait locus (caQTL) mapping using HAECs from 56 donors, allelic imbalance assay from 7 donors, and luciferase assays demonstrate that CAD/IS-protective allele at rs17114036 in PLPP3 intron 5 confers increased endothelial enhancer activity. ChIP-PCR and luciferase assays show that CAD/IS-protective allele at rs17114036 creates a binding site for transcription factor Krüppel-like factor 2 (KLF2), which increases the enhancer activity under unidirectional flow. These results demonstrate that a human SNP contributes to critical endothelial mechanotransduction mechanisms and suggest that human haplotypes and related cis-regulatory elements provide a previously unappreciated layer of regulatory control in cellular mechanosensing mechanisms.
Keywords: GWAS; coronary artery disease; endothelial cells; mechanotransduction; shear stress.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Coronary Artery Disease Risk Variant Dampens the Expression of CALCRL by Reducing HSF Binding to Shear Stress Responsive Enhancer in Endothelial Cells In Vitro.Arterioscler Thromb Vasc Biol. 2024 Jun;44(6):1330-1345. doi: 10.1161/ATVBAHA.123.318964. Epub 2024 Apr 11. Arterioscler Thromb Vasc Biol. 2024. PMID: 38602103 Free PMC article.
-
Mechanosensitive PPAP2B Regulates Endothelial Responses to Atherorelevant Hemodynamic Forces.Circ Res. 2015 Jul 31;117(4):e41-e53. doi: 10.1161/CIRCRESAHA.117.306457. Epub 2015 Jun 1. Circ Res. 2015. PMID: 26034042 Free PMC article.
-
Minor allele C of chromosome 1p32 single nucleotide polymorphism rs11206510 confers risk of ischemic stroke in the Chinese Han population.Stroke. 2010 Aug;41(8):1587-92. doi: 10.1161/STROKEAHA.110.583096. Epub 2010 Jun 24. Stroke. 2010. PMID: 20576952
-
Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants.Stroke. 2014 Jan;45(1):24-36. doi: 10.1161/STROKEAHA.113.002707. Epub 2013 Nov 21. Stroke. 2014. PMID: 24262325 Free PMC article. Review.
-
Mechanosensitive microRNAs-role in endothelial responses to shear stress and redox state.Free Radic Biol Med. 2013 Sep;64:61-8. doi: 10.1016/j.freeradbiomed.2013.05.034. Epub 2013 May 30. Free Radic Biol Med. 2013. PMID: 23727269 Free PMC article. Review.
Cited by
-
Genetic analyses of the electrocardiographic QT interval and its components identify additional loci and pathways.Nat Commun. 2022 Sep 1;13(1):5144. doi: 10.1038/s41467-022-32821-z. Nat Commun. 2022. PMID: 36050321 Free PMC article.
-
Preliminary Study of Genome-Wide Association Identified Novel Susceptibility Genes for Hemorheological Indexes in a Chinese Population.Transfus Med Hemother. 2022 Jul 5;49(6):346-357. doi: 10.1159/000524849. eCollection 2022 Dec. Transfus Med Hemother. 2022. PMID: 36654975 Free PMC article.
-
The contribution of non-coding regulatory elements to cardiovascular disease.Open Biol. 2020 Jul;10(7):200088. doi: 10.1098/rsob.200088. Epub 2020 Jul 1. Open Biol. 2020. PMID: 32603637 Free PMC article. Review.
-
Disentangling the complexity of psoriasis in the post-genome-wide association era.Genes Immun. 2023 Oct;24(5):236-247. doi: 10.1038/s41435-023-00222-x. Epub 2023 Sep 16. Genes Immun. 2023. PMID: 37717118 Review.
-
Genotype inference from aggregated chromatin accessibility data reveals genetic regulatory mechanisms.bioRxiv [Preprint]. 2024 Sep 5:2024.09.04.610850. doi: 10.1101/2024.09.04.610850. bioRxiv. 2024. Update in: Genome Biol. 2025 Mar 30;26(1):81. doi: 10.1186/s13059-025-03538-1. PMID: 39282458 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

