Conformational Differences between Functional Human Immunodeficiency Virus Envelope Glycoprotein Trimers and Stabilized Soluble Trimers
- PMID: 30429345
- PMCID: PMC6340038
- DOI: 10.1128/JVI.01709-18
Conformational Differences between Functional Human Immunodeficiency Virus Envelope Glycoprotein Trimers and Stabilized Soluble Trimers
Abstract
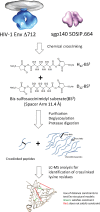
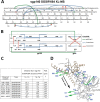
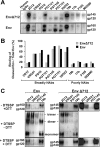
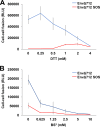
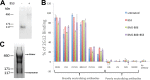
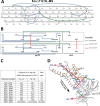
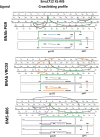
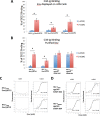
Binding to the receptor CD4 triggers entry-related conformational changes in the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) trimer, (gp120/gp41)3 Soluble versions of HIV-1 Env trimers (sgp140 SOSIP.664) stabilized by a gp120-gp41 disulfide bond and a change (I559P) in gp41 have been structurally characterized. Here, we use cross-linking/mass spectrometry to evaluate the conformations of functional membrane Env and sgp140 SOSIP.664. Differences were detected in the gp120 trimer association domain and C terminus and in the gp41 heptad repeat 1 (HR1) region. Whereas the membrane Env trimer exposes the gp41 HR1 coiled coil only after CD4 binding, the sgp140 SOSIP.664 HR1 coiled coil was accessible to the gp41 HR2 peptide even in the absence of CD4. Our results delineate differences in both gp120 and gp41 subunits between functional membrane Env and the sgp140 SOSIP.664 trimer and provide distance constraints that can assist validation of candidate structural models of the native HIV-1 Env trimer.IMPORTANCE HIV-1 envelope glycoprotein spikes mediate the entry of the virus into host cells and are a major target for vaccine-induced antibodies. Soluble forms of the envelope glycoproteins that are stable and easily produced have been characterized extensively and are being considered as vaccines. Here, we present evidence that these stabilized soluble envelope glycoproteins differ in multiple respects from the natural HIV-1 envelope glycoproteins. By pinpointing these differences, our results can guide the improvement of envelope glycoprotein preparations to achieve greater similarity to the viral envelope glycoprotein spike, potentially increasing their effectiveness as a vaccine.
Keywords: conformation; cross-linker; envelope glycoprotein; human immunodeficiency virus; mass spectrometry; stabilization; structure; trimer.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
SOSIP Changes Affect Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Conformation and CD4 Engagement.J Virol. 2018 Sep 12;92(19):e01080-18. doi: 10.1128/JVI.01080-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021898 Free PMC article.
-
Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer.PLoS One. 2015 Apr 7;10(4):e0122111. doi: 10.1371/journal.pone.0122111. eCollection 2015. PLoS One. 2015. PMID: 25849367 Free PMC article.
-
HIV-1 gp41 Residues Modulate CD4-Induced Conformational Changes in the Envelope Glycoprotein and Evolution of a Relaxed Conformation of gp120.J Virol. 2018 Jul 31;92(16):e00583-18. doi: 10.1128/JVI.00583-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875245 Free PMC article.
-
HIV-1 envelope glycoprotein structure.Curr Opin Struct Biol. 2013 Apr;23(2):268-76. doi: 10.1016/j.sbi.2013.03.007. Epub 2013 Apr 18. Curr Opin Struct Biol. 2013. PMID: 23602427 Free PMC article. Review.
-
Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
Cited by
-
Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET.Nature. 2019 Apr;568(7752):415-419. doi: 10.1038/s41586-019-1101-y. Epub 2019 Apr 10. Nature. 2019. PMID: 30971821 Free PMC article.
-
Inhibition of human immunodeficiency virus (HIV-1) infectivity by expression of poorly or broadly neutralizing antibodies against Env in virus-producing cells.J Virol. 2024 Feb 20;98(2):e0159423. doi: 10.1128/jvi.01594-23. Epub 2024 Jan 30. J Virol. 2024. PMID: 38289101 Free PMC article.
-
SOS and IP Modifications Predominantly Affect the Yield but Not Other Properties of SOSIP.664 HIV-1 Env Glycoprotein Trimers.J Virol. 2019 Dec 12;94(1):e01521-19. doi: 10.1128/JVI.01521-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31619555 Free PMC article.
-
Shedding-Resistant HIV-1 Envelope Glycoproteins Adopt Downstream Conformations That Remain Responsive to Conformation-Preferring Ligands.J Virol. 2020 Aug 17;94(17):e00597-20. doi: 10.1128/JVI.00597-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32522853 Free PMC article.
-
Single-Molecule FRET Imaging of Virus Spike-Host Interactions.Viruses. 2021 Feb 21;13(2):332. doi: 10.3390/v13020332. Viruses. 2021. PMID: 33669922 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

