Epstein-Barr Virus Infection of Cell Lines Derived from Diffuse Large B-Cell Lymphomas Alters MicroRNA Loading of the Ago2 Complex
- PMID: 30429351
- PMCID: PMC6340033
- DOI: 10.1128/JVI.01297-18
Epstein-Barr Virus Infection of Cell Lines Derived from Diffuse Large B-Cell Lymphomas Alters MicroRNA Loading of the Ago2 Complex
Abstract
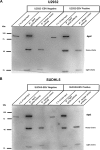
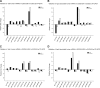
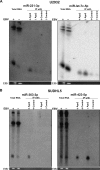
Diffuse large B-cell lymphoma (DLBCL) is an aggressive lymphoid tumor which is occasionally Epstein-Barr virus (EBV) positive and is further subtyped as activated B-cell DLBCL (ABC-DLBCL) and germinal center B-cell DLBCL (GCB-DLBCL), which has implications for prognosis and treatment. We performed Ago2 RNA immunoprecipitation followed by high-throughput RNA sequencing (Ago2-RIP-seq) to capture functionally active microRNAs (miRNAs) in EBV-negative ABC-DLBCL and GCB-DLBCL cell lines and their EBV-infected counterparts. In parallel, total miRNA profiles of these cells were determined to capture the cellular miRNA profile for comparison with the functionally active profile. Selected miRNAs with differential abundances were validated using real-time quantitative PCR (RT-qPCR) and Northern blotting. We found 6 miRNAs with differential abundances (2 upregulated and 4 downregulated miRNAs) between EBV-negative and -positive ABC-DLBCL cells and 12 miRNAs with differential abundances (3 upregulated and 9 downregulated miRNAs) between EBV-negative and -positive GCB-DLBCL cells. Eight and twelve miRNAs were confirmed using RT-qPCR in ABC-DLBCL and GCB-DLBCL cells, respectively. Selected miRNAs were analyzed in additional type I/II versus type III EBV latency DLBCL cell lines. Furthermore, upregulation of miR-221-3p and downregulation of let7c-5p in ABC-DLBCL cells and upregulation of miR-363-3p and downregulation of miR-423-5p in GCB-DLBCL cells were verified using RIP-Northern blotting. Our comprehensive sequence analysis of the DLBCL miRNA profiles identified sets of deregulated miRNAs by Ago2-RIP-seq. Our Ago2-IP-seq miRNA profile could be considered an important data set for the detection of deregulated functionally active miRNAs in DLBCLs and could possibly lead to the identification of miRNAs as biomarkers for the classification of DLBCLs or even as targets for personalized targeted treatment.IMPORTANCE Diffuse large B-cell lymphoma (DLBCL) is a highly aggressive tumor of lymphoid origin which is occasionally Epstein-Barr virus (EBV) positive. MicroRNAs are found in most multicellular organisms and even in viruses such as EBV. They regulate the synthesis of proteins by binding to their cognate mRNA. MicroRNAs are tethered to their target mRNAs by "Argonaute" proteins. Here we compared the overall miRNA content of the Ago2 complex by differential loading to the overall content of miRNAs in two DLBCL cell lines and their EBV-converted counterparts. In all cell lines, the Ago2 load was different from the overall expression of miRNAs. In addition, the loading of the Ago2 complex was changed upon infection with EBV. This indicates that the virus not only changes the overall content of miRNAs but also influences the expression of proteins by affecting the Ago complexes.
Keywords: Ago-IP; Argonaute; DLBCL; Epstein-Barr virus; microRNA; profiling; sequencing.
Copyright © 2019 American Society for Microbiology.
Figures
References
-
- Rickinson AB, Kieff E. 2007. Epstein-Barr virus, p 2655–2700. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Strauss SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

