Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer
- PMID: 30429477
- PMCID: PMC6235880
- DOI: 10.1038/s41467-018-07041-z
Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer
Abstract
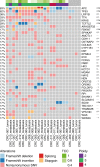
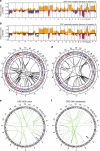
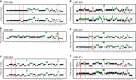
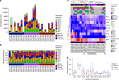
Incomplete understanding of the metastatic process hinders personalized therapy. Here we report the most comprehensive whole-genome study of colorectal metastases vs. matched primary tumors. 65% of somatic mutations originate from a common progenitor, with 15% being tumor- and 19% metastasis-specific, implicating a higher mutation rate in metastases. Tumor- and metastasis-specific mutations harbor elevated levels of BRCAness. We confirm multistage progression with new components ARHGEF7/ARHGEF33. Recurrently mutated non-coding elements include ncRNAs RP11-594N15.3, AC010091, SNHG14, 3' UTRs of FOXP2, DACH2, TRPM3, XKR4, ANO5, CBL, CBLB, the latter four potentially dual protagonists in metastasis and efferocytosis-/PD-L1 mediated immunosuppression. Actionable metastasis-specific lesions include FAT1, FGF1, BRCA2, KDR, and AKT2-, AKT3-, and PDGFRA-3' UTRs. Metastasis specific mutations are enriched in PI3K-Akt signaling, cell adhesion, ECM and hepatic stellate activation genes, suggesting genetic programs for site-specific colonization. Our results put forward hypotheses on tumor and metastasis evolution, and evidence for metastasis-specific events relevant for personalized therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

