Pathway analysis of a genome‑wide association study on a long non‑coding RNA expression profile in oral squamous cell carcinoma
- PMID: 30431131
- PMCID: PMC6312939
- DOI: 10.3892/or.2018.6870
Pathway analysis of a genome‑wide association study on a long non‑coding RNA expression profile in oral squamous cell carcinoma
Abstract
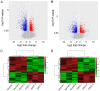
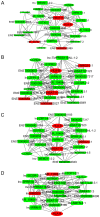
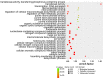
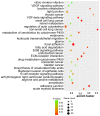
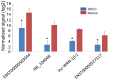
Long non‑coding RNAs (lncRNAs) have been consistently demonstrated to be involved in oral squamous cell carcinoma (OSCC) as either tumor oncogenes or tumor suppressors. However, the underlying mechanisms of OSCC tumorigenesis and development have not yet been fully elucidated. The expression profiles of mRNAs and lncRNAs in OSCC were analyzed by a microarray assay. To verify the results of the microarray, 10 differentially expressed lncRNAs were randomly selected and measured by quantitative RT‑PCR (qRT‑PCR). Gene Ontology (GO) and metabolic pathway analyses were performed to analyze gene function and identify enriched pathways. Subsequently, two independent algorithms were used to predict the target genes of the lncRNAs. We identified 2,294 lncRNAs and 1,938 mRNAs that were differentially expressed in all three OSCC tissues by a microarray assay. Through the construction of co‑expression networks of differentially expressed genes, 4 critical lncRNAs nodes were identified as potential key factors in the pathogenesis of OSCC. Expression of the 4 critical lncRNA nodes was not associated with age, sex, smoking or tumor location (P>0.05) but was positively correlated with clinical stage, lymphatic metastasis, distant metastasis and survival status (P<0.05). Kaplan‑Meier analysis demonstrated that low expression levels of these 4 critical lncRNA nodes contributed to poor median progression‑free survival (PFS) and overall survival (OS) (P<0.05). GO and pathway analyses indicated that the functions and enriched pathways of many dysregulated genes are associated with cancer. Potential target genes of dysregulated lncRNAs were enriched in 43 metabolic pathways, with cancer pathways being the primary enrichment pathways. In summary, we analyzed the profile of lncRNAs in OSCC and identified the functions and enriched metabolic pathways of both dysregulated mRNAs and the target genes of dysregulated lncRNAs, providing new insights into molecular markers and therapeutic targets for OSCC.
Figures





Similar articles
-
Potential role of differentially expressed lncRNAs in the pathogenesis of oral squamous cell carcinoma.Arch Oral Biol. 2015 Oct;60(10):1581-7. doi: 10.1016/j.archoralbio.2015.08.003. Epub 2015 Aug 4. Arch Oral Biol. 2015. PMID: 26276270
-
Long noncoding RNA MYOSLID promotes invasion and metastasis by modulating the partial epithelial-mesenchymal transition program in head and neck squamous cell carcinoma.J Exp Clin Cancer Res. 2019 Jun 25;38(1):278. doi: 10.1186/s13046-019-1254-4. J Exp Clin Cancer Res. 2019. PMID: 31238980 Free PMC article.
-
Exploring the long noncoding RNAs-based biomarkers and pathogenesis of malignant transformation from dysplasia to oral squamous cell carcinoma by bioinformatics method.Eur J Cancer Prev. 2020 Mar;29(2):174-181. doi: 10.1097/CEJ.0000000000000527. Eur J Cancer Prev. 2020. PMID: 31343435 Free PMC article.
-
Novel insights on oral squamous cell carcinoma management using long non-coding RNAs.Oncol Res. 2024 Sep 18;32(10):1589-1612. doi: 10.32604/or.2024.052120. eCollection 2024. Oncol Res. 2024. PMID: 39308526 Free PMC article. Review.
-
Long non-coding RNAs in Oral squamous cell carcinoma: biologic function, mechanisms and clinical implications.Mol Cancer. 2019 May 27;18(1):102. doi: 10.1186/s12943-019-1021-3. Mol Cancer. 2019. PMID: 31133028 Free PMC article. Review.
Cited by
-
LncRNA EIF3J-DT promotes chemoresistance in oral squamous cell carcinoma.Oral Dis. 2024 Nov;30(8):4909-4920. doi: 10.1111/odi.14987. Epub 2024 May 30. Oral Dis. 2024. PMID: 38817073 Free PMC article.
-
Long non‑coding RNAs are novel players in oral inflammatory disorders, potentially premalignant oral epithelial lesions and oral squamous cell carcinoma (Review).Int J Mol Med. 2020 Aug;46(2):535-545. doi: 10.3892/ijmm.2020.4628. Epub 2020 Jun 3. Int J Mol Med. 2020. PMID: 32626947 Free PMC article. Review.
-
Elevated RUNX1 is a prognostic biomarker for human head and neck squamous cell carcinoma.Exp Biol Med (Maywood). 2021 Mar;246(5):538-546. doi: 10.1177/1535370220969663. Epub 2020 Nov 26. Exp Biol Med (Maywood). 2021. PMID: 33241710 Free PMC article.
-
The Emerging Role of Exosomes in Oral Squamous Cell Carcinoma.Front Cell Dev Biol. 2021 Feb 22;9:628103. doi: 10.3389/fcell.2021.628103. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33718365 Free PMC article. Review.
-
Circulating Long Non-Coding RNAs Could Be the Potential Prognostic Biomarker for Liquid Biopsy for the Clinical Management of Oral Squamous Cell Carcinoma.Cancers (Basel). 2022 Nov 14;14(22):5590. doi: 10.3390/cancers14225590. Cancers (Basel). 2022. PMID: 36428681 Free PMC article. Review.
References
-
- Cui Z, Ren S, Lu J, Wang F, Xu W, Sun Y, Wei M, Chen J, Gao X, Xu C, et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol. 2013;31:1117–1123. doi: 10.1016/j.urolonc.2011.11.030. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

