Broadly Neutralizing Antibody Mediated Clearance of Human Hepatitis C Virus Infection
- PMID: 30439341
- PMCID: PMC6250073
- DOI: 10.1016/j.chom.2018.10.012
Broadly Neutralizing Antibody Mediated Clearance of Human Hepatitis C Virus Infection
Abstract
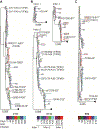
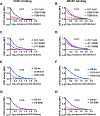
The role that broadly neutralizing antibodies (bNAbs) play in natural clearance of human hepatitis C virus (HCV) infection and the underlying mechanisms remain unknown. Here, we investigate the mechanism by which bNAbs, isolated from two humans who spontaneously cleared HCV infection, contribute to HCV control. Using viral gene sequences amplified from longitudinal plasma of the two subjects, we found that these bNAbs, which target the front layer of the HCV envelope protein E2, neutralized most autologous HCV strains. Acquisition of resistance to bNAbs by some autologous strains was accompanied by progressive loss of E2 protein function, and temporally associated with HCV clearance. These data demonstrate that bNAbs can mediate clearance of human HCV infection by neutralizing infecting strains and driving escaped viruses to an unfit state. These immunopathologic events distinguish HCV from HIV-1 and suggest that development of an HCV vaccine may be achievable.
Keywords: HCV clearance; antibody specificity; binding sites; hepatitis C virus; immunologic memory; neutralizing antibodies.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
A.I.F., J.E.C., G.M.S., and J.R.B. are inventors of patents submitted pertaining to some of the antibodies and antigens presented in this paper. J.E.C. has served as a consultant for Takeda Vaccines, Sanofi Pasteur, Pfizer, and Novavax, is on the Scientific Advisory Boards of CompuVax, GigaGen, Meissa Vaccines, PaxVax, and is Founder of IDBiologics, Inc. The other authors declare no competing interests.
Figures





Comment in
-
Hepatitis C Virus Neutralizing Antibodies: Is a Vaccine Still Possible?Cell Host Microbe. 2018 Nov 14;24(5):620-621. doi: 10.1016/j.chom.2018.10.017. Cell Host Microbe. 2018. PMID: 30439338
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

