Karyotype heterogeneity in Philodendron s.l. (Araceae) revealed by chromosome mapping of rDNA loci
- PMID: 30440003
- PMCID: PMC6237374
- DOI: 10.1371/journal.pone.0207318
Karyotype heterogeneity in Philodendron s.l. (Araceae) revealed by chromosome mapping of rDNA loci
Abstract
Philodendron s.l. (Araceae) has been recently focus of taxonomic and phylogenetic studies, but karyotypic data are limited to chromosome numbers and a few published genome sizes. In this work, karyotypes of 34 species of Philodendron s.l. (29 species of Philodendron and five of Thaumatophyllum), ranging from 2n = 28 to 36 chromosomes, were analyzed by fluorescence in situ hybridization (FISH) with rDNA and telomeric probes, aiming to understand the evolution of the karyotype diversity of the group. Philodendron presented a high number variation of 35S rDNA, ranging from two to 16 sites, which were mostly in the terminal region of the short arms, with nine species presenting heteromorphisms. In the case of Thaumatophyllum species, we observed a considerably lower variation, which ranged from two to four terminal sites. The distribution of the 5S rDNA clusters was more conserved, with two sites for most species, being preferably located interstitially in the long chromosome arms. For the telomeric probe, while exclusively terminal sites were observed for P. giganteum (2n = 30) chromosomes, P. callosum (2n = 28) presented an interstitial distribution associated with satellite DNA. rDNA sites of the analyzed species of Philodendron s.l. species were randomly distributed considering the phylogenetic context, probably due to rapid evolution and great diversity of these genomes. The observed heteromorphisms suggest the accumulation of repetitive DNA in the genomes of some species and the occurrence of chromosomal rearrangements along the karyotype evolution of the group.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
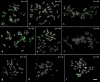
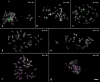



Similar articles
-
Cytogenetic features of rRNA genes across land plants: analysis of the Plant rDNA database.Plant J. 2017 Mar;89(5):1020-1030. doi: 10.1111/tpj.13442. Epub 2017 Feb 14. Plant J. 2017. PMID: 27943584
-
rDNA Loci Evolution in the Genus Glechoma (Lamiaceae).PLoS One. 2016 Nov 21;11(11):e0167177. doi: 10.1371/journal.pone.0167177. eCollection 2016. PLoS One. 2016. PMID: 27870903 Free PMC article.
-
Karyotype analysis of eight cultivated Allium species.J Appl Genet. 2019 Feb;60(1):1-11. doi: 10.1007/s13353-018-0474-1. Epub 2018 Oct 23. J Appl Genet. 2019. PMID: 30353472 Free PMC article.
-
Interstitial telomeric sites and Robertsonian translocations in species of Ipheion and Nothoscordum (Amaryllidaceae).Genetica. 2016 Apr;144(2):157-66. doi: 10.1007/s10709-016-9886-1. Epub 2016 Feb 11. Genetica. 2016. PMID: 26869260 Review.
-
A multidisciplinary and integrative review of the structural genome and epigenome of Capsicum L. species.Planta. 2025 Mar 9;261(4):82. doi: 10.1007/s00425-025-04653-w. Planta. 2025. PMID: 40057910 Review.
Cited by
-
Distinguishing Sichuan Walnut Cultivars and Examining Their Relationships with Juglans regia and J. sigillata by FISH, Early-Fruiting Gene Analysis, and SSR Analysis.Front Plant Sci. 2020 Feb 25;11:27. doi: 10.3389/fpls.2020.00027. eCollection 2020. Front Plant Sci. 2020. PMID: 32161605 Free PMC article.
-
The Use of Ribosomal DNA for Comparative Cytogenetics.Methods Mol Biol. 2023;2672:265-284. doi: 10.1007/978-1-0716-3226-0_17. Methods Mol Biol. 2023. PMID: 37335483
-
Molecular and Cytogenetic Analysis of rDNA Evolution in Crepis Sensu Lato.Int J Mol Sci. 2022 Mar 26;23(7):3643. doi: 10.3390/ijms23073643. Int J Mol Sci. 2022. PMID: 35409003 Free PMC article.
-
Fluorescence In Situ Hybridization (FISH) Analysis of the Locations of the Oligonucleotides 5S rDNA, (AGGGTTT)3, and (TTG)6 in Three Genera of Oleaceae and Their Phylogenetic Framework.Genes (Basel). 2019 May 17;10(5):375. doi: 10.3390/genes10050375. Genes (Basel). 2019. PMID: 31108932 Free PMC article.
-
Genetic relationship study of some Vicia species by FISH and total seed storage protein patterns.J Genet Eng Biotechnol. 2020 Jul 31;18(1):37. doi: 10.1186/s43141-020-00054-6. J Genet Eng Biotechnol. 2020. PMID: 32737692 Free PMC article.
References
-
- Mayo SJ, Bogner J, Boyce PC. The genera of Araceae. Richmond: Royal Botanic Gardens, Kew, 1997; 370 p. 10.2307/4110410 - DOI
-
- Boyce PC, Croat TB. The Überlist of Araceae, totals for published and estimated number of species in aroid genera. 2018. http://www.aroid.org/genera/180211uberlist.pdf 11 Feb 2018.
-
- Calazans LSB, Sakuragui CM, Mayo SJ. From open areas to forests? The evolutionary history of Philodendron subgenus Meconostigma (Araceae) using morphological data. Flora. 2014;209: 117–121. 10.1016/j.flora.2013.12.004 - DOI
-
- Loss-Oliveira L, Sakuragui C, Soares ML, Schrago CG. Evolution of Philodendron (Araceae) species in Neotropical biomes. PeerJ. 2016;4: e1744 10.7717/peerj.1744 - DOI - PMC - PubMed
-
- Mayo SJ. A revision of Philodendron subgenus Meconostigma (Araceae). Kew Bulletin. 1991;46: 601–681.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

