Phylotranscriptomics suggests the jawed vertebrate ancestor could generate diverse helper and regulatory T cell subsets
- PMID: 30442091
- PMCID: PMC6238376
- DOI: 10.1186/s12862-018-1290-2
Phylotranscriptomics suggests the jawed vertebrate ancestor could generate diverse helper and regulatory T cell subsets
Abstract
Background: The cartilaginous fishes diverged from other jawed vertebrates ~ 450 million years ago (mya). Despite this key evolutionary position, the only high-quality cartilaginous fish genome available is for the elephant shark (Callorhinchus milii), a chimaera whose ancestors split from the elasmobranch lineage ~ 420 mya. Initial analysis of this resource led to proposals that key components of the cartilaginous fish adaptive immune system, most notably their array of T cell subsets, was primitive compared to mammals. This proposal is at odds with the robust, antigen-specific antibody responses reported in elasmobranchs following immunization. To explore this discrepancy, we generated a multi-tissue transcriptome for small-spotted catshark (Scyliorhinus canicula), a tractable elasmobranch model for functional studies. We searched this, and other newly available sequence datasets, for CD4+ T cell subset-defining genes, aiming to confirm the presence or absence of each subset in cartilaginous fishes.
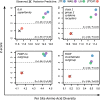
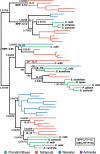
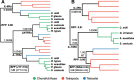
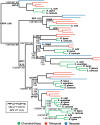
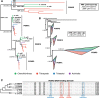
Results: We generated a new transcriptome based on a normalised, multi-tissue RNA pool, aiming to maximise representation of tissue-specific and lowly expressed genes. We utilized multiple transcriptomic datasets and assembly variants in phylogenetic reconstructions to unambiguously identify several T cell subset-specific molecules in cartilaginous fishes for the first time, including interleukins, interleukin receptors, and key transcription factors. Our results reveal the inability of standard phylogenetic reconstruction approaches to capture the site-specific evolutionary processes of fast-evolving immune genes but show that site-heterogeneous mixture models can adequately do so.
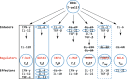
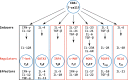
Conclusions: Our analyses reveal that cartilaginous fishes are capable of producing a range of CD4+ T cell subsets comparable to that of mammals. Further, that the key molecules required for the differentiation and functioning of these subsets existed in the jawed vertebrate ancestor. Additionally, we highlight the importance of considering phylogenetic diversity and, where possible, utilizing multiple datasets for individual species, to accurately infer gene presence or absence at higher taxonomic levels.
Keywords: Cartilaginous fish; Elasmobranch; Immune gene evolution; Immunity; Interleukin; Phylogenetic rooting; Shark; Site-heterogeneous models; T cell; Transcriptome.
Conflict of interest statement
Ethics approval and consent to participate
All animal procedures were conducted in accordance with UK Home Office ‘Animals and Scientific Procedures Act 1986; Amendment Regulations 2012’ on animal care and use, with prior ethical approval from the University of Aberdeen’s Animal Welfare and Ethical Review Body (AWERB).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Dooley H. Athena and the evolution of adaptive immunity. In: Smith S, Sim B, Flajnik MF, editors. Chapter 2 in Immunobiology of the Shark, Taylor and Francis Group, LLC. 2014. p. 24-50.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

