CNBP controls IL-12 gene transcription and Th1 immunity
- PMID: 30442645
- PMCID: PMC6279399
- DOI: 10.1084/jem.20181031
CNBP controls IL-12 gene transcription and Th1 immunity
Abstract
An inducible program of inflammatory gene expression is a hallmark of antimicrobial defenses. Recently, cellular nucleic acid-binding protein (CNBP) was identified as a regulator of nuclear factor-kappaB (NF-κB)-dependent proinflammatory cytokine gene expression. Here, we generated mice lacking CNBP and found that CNBP regulates a very restricted gene signature that includes IL-12β. CNBP resides in the cytosol of macrophages and translocates to the nucleus in response to diverse microbial pathogens and pathogen-derived products. Cnbp-deficient macrophages induced canonical NF-κB/Rel signaling normally but were impaired in their ability to control the activation of c-Rel, a key driver of IL-12β gene transcription. The nuclear translocation and DNA-binding activity of c-Rel required CNBP. Lastly, Cnbp-deficient mice were more susceptible to acute toxoplasmosis associated with reduced production of IL-12β, as well as a reduced T helper type 1 (Th1) cell IFN-γ response essential to controlling parasite replication. Collectively, these findings identify CNBP as important regulator of c-Rel-dependent IL-12β gene transcription and Th1 immunity.
© 2018 Chen et al.
Figures

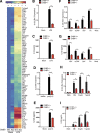
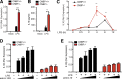

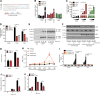

Comment in
-
Sensing danger through a "finger".J Exp Med. 2018 Dec 3;215(12):2969-2971. doi: 10.1084/jem.20182034. Epub 2018 Nov 20. J Exp Med. 2018. PMID: 30459157 Free PMC article.
Similar articles
-
A proximal kappaB site in the IL-23 p19 promoter is responsible for RelA- and c-Rel-dependent transcription.J Immunol. 2007 Nov 15;179(10):6596-603. doi: 10.4049/jimmunol.179.10.6596. J Immunol. 2007. PMID: 17982049
-
Deficient translocation of c-Rel is associated with impaired Th1 cytokine production in T cells from atopic dermatitis patients.Exp Dermatol. 2005 Jan;14(1):17-25. doi: 10.1111/j.0906-6705.2005.00241.x. Exp Dermatol. 2005. PMID: 15660915
-
Involvement of Notch signaling pathway in regulating IL-12 expression via c-Rel in activated macrophages.Mol Immunol. 2012 Jul;51(3-4):255-62. doi: 10.1016/j.molimm.2012.03.017. Epub 2012 Mar 29. Mol Immunol. 2012. PMID: 22463790 Free PMC article.
-
Distinctions between c-Rel and other NF-kappaB proteins in immunity and disease.Bioessays. 2003 Aug;25(8):767-80. doi: 10.1002/bies.10306. Bioessays. 2003. PMID: 12879447 Review.
-
What's new about CNBP? Divergent functions and activities for a conserved nucleic acid binding protein.Biochim Biophys Acta Gen Subj. 2021 Nov;1865(11):129996. doi: 10.1016/j.bbagen.2021.129996. Epub 2021 Aug 30. Biochim Biophys Acta Gen Subj. 2021. PMID: 34474118 Review.
Cited by
-
Cellular nucleic acid-binding protein is essential for type I interferon-mediated immunity to RNA virus infection.Proc Natl Acad Sci U S A. 2021 Jun 29;118(26):e2100383118. doi: 10.1073/pnas.2100383118. Proc Natl Acad Sci U S A. 2021. PMID: 34168080 Free PMC article.
-
Caspase-8 promotes c-Rel-dependent inflammatory cytokine expression and resistance against Toxoplasma gondii.Proc Natl Acad Sci U S A. 2019 Jun 11;116(24):11926-11935. doi: 10.1073/pnas.1820529116. Epub 2019 May 30. Proc Natl Acad Sci U S A. 2019. PMID: 31147458 Free PMC article.
-
Development of Therapeutic Approaches for Myotonic Dystrophies Type 1 and Type 2.Int J Mol Sci. 2022 Sep 10;23(18):10491. doi: 10.3390/ijms231810491. Int J Mol Sci. 2022. PMID: 36142405 Free PMC article. Review.
-
Single-cell transcriptome profiling reveals enriched memory T-cell subpopulations in hypertension.Front Cell Dev Biol. 2023 Mar 16;11:1132040. doi: 10.3389/fcell.2023.1132040. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37009484 Free PMC article.
-
Toll-like Receptors and the Control of Immunity.Cell. 2020 Mar 19;180(6):1044-1066. doi: 10.1016/j.cell.2020.02.041. Epub 2020 Mar 11. Cell. 2020. PMID: 32164908 Free PMC article. Review.
References
-
- Andrade W.A., Souza M.C., Ramos-Martinez E., Nagpal K., Dutra M.S., Melo M.B., Bartholomeu D.C., Ghosh S., Golenbock D.T., and Gazzinelli R.T.. 2013. Combined action of nucleic acid-sensing Toll-like receptors and TLR11/TLR12 heterodimers imparts resistance to Toxoplasma gondii in mice. Cell Host Microbe. 13:42–53. 10.1016/j.chom.2012.12.003 - DOI - PMC - PubMed
-
- Aste-Amezaga M., Ma X., Sartori A., and Trinchieri G.. 1998. Molecular mechanisms of the induction of IL-12 and its inhibition by IL-10. J. Immunol. 160:5936–5944. - PubMed
-
- Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J., et al. . 2016. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 165:1672–1685. 10.1016/j.cell.2016.05.075 - DOI - PMC - PubMed
-
- Benhalevy D., Gupta S.K., Danan C.H., Ghosal S., Sun H.W., Kazemier H.G., Paeschke K., Hafner M., and Juranek S.A.. 2017. The Human CCHC-type Zinc Finger Nucleic Acid-Binding Protein Binds G-Rich Elements in Target mRNA Coding Sequences and Promotes Translation. Cell Reports. 18:2979–2990. 10.1016/j.celrep.2017.02.080 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

