Structure of human TFIID and mechanism of TBP loading onto promoter DNA
- PMID: 30442764
- PMCID: PMC6446905
- DOI: 10.1126/science.aau8872
Structure of human TFIID and mechanism of TBP loading onto promoter DNA
Abstract
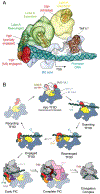
The general transcription factor IID (TFIID) is a critical component of the eukaryotic transcription preinitiation complex (PIC) and is responsible for recognizing the core promoter DNA and initiating PIC assembly. We used cryo-electron microscopy, chemical cross-linking mass spectrometry, and biochemical reconstitution to determine the complete molecular architecture of TFIID and define the conformational landscape of TFIID in the process of TATA box-binding protein (TBP) loading onto promoter DNA. Our structural analysis revealed five structural states of TFIID in the presence of TFIIA and promoter DNA, showing that the initial binding of TFIID to the downstream promoter positions the upstream DNA and facilitates scanning of TBP for a TATA box and the subsequent engagement of the promoter. Our findings provide a mechanistic model for the specific loading of TBP by TFIID onto the promoter.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




Comment in
-
IID in 3D: Improved Resolution of Transcription Factor Structure by Cryo-Electron Microscopy.Biochemistry. 2019 Jun 11;58(23):2653-2654. doi: 10.1021/acs.biochem.9b00102. Epub 2019 May 31. Biochemistry. 2019. PMID: 31150224 Free PMC article. No abstract available.
References
-
- Reinberg D, Horikoshi M, Roeder RG, Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J. Biol. Chem 262,3322–3330 (1987). - PubMed

