LZTR1 is a regulator of RAS ubiquitination and signaling
- PMID: 30442766
- PMCID: PMC6794158
- DOI: 10.1126/science.aap8210
LZTR1 is a regulator of RAS ubiquitination and signaling
Abstract
In genetic screens aimed at understanding drug resistance mechanisms in chronic myeloid leukemia cells, inactivation of the cullin 3 adapter protein-encoding leucine zipper-like transcription regulator 1 (LZTR1) gene led to enhanced mitogen-activated protein kinase (MAPK) pathway activity and reduced sensitivity to tyrosine kinase inhibitors. Knockdown of the Drosophila LZTR1 ortholog CG3711 resulted in a Ras-dependent gain-of-function phenotype. Endogenous human LZTR1 associates with the main RAS isoforms. Inactivation of LZTR1 led to decreased ubiquitination and enhanced plasma membrane localization of endogenous KRAS (V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog). We propose that LZTR1 acts as a conserved regulator of RAS ubiquitination and MAPK pathway activation. Because LZTR1 disease mutations failed to revert loss-of-function phenotypes, our findings provide a molecular rationale for LZTR1 involvement in a variety of inherited and acquired human disorders.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
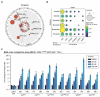
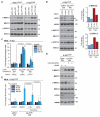


Comment in
-
Ras tuning.Nat Chem Biol. 2019 Feb;15(2):95. doi: 10.1038/s41589-018-0219-9. Nat Chem Biol. 2019. PMID: 30659296 No abstract available.
References
-
- Ren R. Mechanisms of BCR–ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. - PubMed
-
- O'Hare T, Zabriskie MS, Eiring AM, Deininger MW. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat Rev Cancer. 2012;12:513–526. - PubMed
-
- Yamamoto GL, et al. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J Med Genet. 2015;52:413–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

