Study of expression analysis of SIRT4 and the coordinate regulation of bovine adipocyte differentiation by SIRT4 and its transcription factors
- PMID: 30442871
- PMCID: PMC6294651
- DOI: 10.1042/BSR20181705
Study of expression analysis of SIRT4 and the coordinate regulation of bovine adipocyte differentiation by SIRT4 and its transcription factors
Abstract
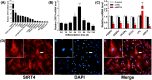
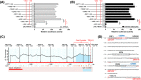
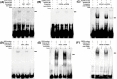
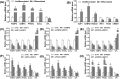
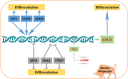
Sirtuins, NAD+-dependent deacylases and ADP-ribosyltransferases, are critical regulators of metabolism involved in many biological processes, and are involved in mediating adaptive responses to the cellular environment. SIRT4 is a mitochondrial sirtuin and has been shown to play a critical role in maintaining insulin secretion and glucose homeostasis. As a regulator of lipid homeostasis, SIRT4 can repress fatty acid oxidation and promote lipid anabolism in nutrient-replete conditions. Using real-time quantitative PCR (qPCR) to explore the molecular mechanisms of transcriptional regulation of bovine SIRT4 during adipocyte differentiation, we found that bovine SIRT4 is expressed at high levels in bovine subcutaneous adipose tissue. SIRT4 knockdown led to decreased expression of adipogenic differentiation marker genes during adipocyte differentiation. The core promoter of bovine SIRT4 was identified in the -402/-60 bp region of the cloned 2-kb fragment containing the 5'-regulatory region. Binding sites were identified in this region for E2F transcription factor-1 (E2F1), CCAAT/enhancer-binding protein β (CEBPβ), homeobox A5 (HOXA5), interferon regulatory factor 4 (IRF4), paired box 4 (PAX4), and cAMP responsive element-binding protein 1 (CREB1) by using Electrophoretic mobility shift assay (EMSA) and luciferase reporter gene assay. We also found that E2F1, CEBPβ, and HOXA5 transcriptionally activate SIRT4 expression, whereas, IRF4, PAX4, and CREB1 transcriptionally repress SIRT4 expression. We further verified that SIRT4 knockdown could affect the ability of these transcription factors (TFs) to regulate the differentiation of bovine adipocytes. In conclusion, our results shed light on the mechanisms underlying the transcriptional regulation of SIRT4 expression in bovine adipocytes.
Keywords: SIRT4; adipocyte; bovine; promoter region; transcription factor; transcription regulation.
© 2018 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

