MiR-384 induces apoptosis and autophagy of non-small cell lung cancer cells through the negative regulation of Collagen α-1(X) chain gene
- PMID: 30442874
- PMCID: PMC6356039
- DOI: 10.1042/BSR20181523
MiR-384 induces apoptosis and autophagy of non-small cell lung cancer cells through the negative regulation of Collagen α-1(X) chain gene
Abstract
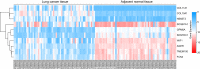
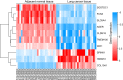
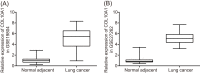
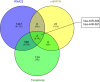
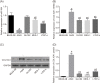
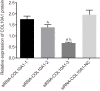
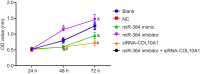
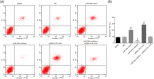
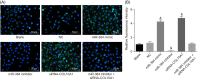
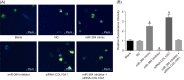
The present study aims to investigate the mechanism of miR-384 in non-small cell lung cancer (NSCLC) cell apoptosis and autophagy by regulating Collagen α-1(X) chain (COL10A1). Bioinformatics methods were applied to evaluate potential miRNAs and genes that might correlate with NSCLC. Tumor tissues and adjacent tissues from 104 NSCLC patients were collected and human NSCLC A549 cell line was selected for subsequent experiments. A549 cells were treated with miR-384 mimic, miR-384 inhibitor, or knockdown of COL10A1. Quantitative real-time PCR (qRT-PCR) and Western blotting were utilized to detect the levels of miR-384, COL10A, Survivin, Bcl-2, Bax, Bcl-xl, Beclin 1, and LC3 in tissues and cells. A series of biological assays including MTT assay, Annexin V-FITC/PI (propidium iodide) staining, immunofluorescence, monodansylcadaverine (MDC) staining were conducted to investigate the effects of miR-384 and COL10A1 on NSCLC cells. Tumorigenicity assay for nude rats was applied. Results obtained from the present study indicated that miR-384 down-regulated COL10A1 by targetting it. Compared with adjacent tissues, miR-384 expression was obviously reduced while COL10A1 expression was significantly enhanced in NSCLC tissues (all P<0.05). Outcomes in vivo and in vitro suggested that cell proliferation and tumorigenicity were inhibited while cell apoptosis and autophagy were induced in NSCLC cells treated with up-regulation of miR-384 or silence of COL10A1. In miR-384 inhibitor group, cell proliferation was improved, while cell apoptosis was reduced and cell autophagy was decreased whereas tumorigenicity of cells was strengthened. Based on the findings of our study, it was established that miR-384 could down-regulate COL10A1 levels, subsequently inhibiting cell proliferation and promoting cell apoptosis and autophagy in NSCLC cells.
Keywords: COL10A1; Cell apoptosis; Cell autophagy; Non-small cell lung cancer; miR-384.
© 2019 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures





References
-
- Lin T.C., Lin P.L., Cheng Y.W., Wu T.C., Chou M.C., Chen C.Y.. et al. (2015) MicroRNA-184 deregulated by the microRNA-21 promotes tumor malignancy and poor outcomes in non-small cell lung cancer via targeting CDC25A and c-Myc. Ann. Surg. Oncol. 22, S1532–S1539 10.1245/s10434-015-4595-z - DOI - PubMed
-
- Wolff H.B., Alberts L., Kastelijn E.A., Lissenberg-Witte B.I., Twisk J.W., Lagerwaard F.J.. et al. (2018) Differences in longitudinal health utility between stereotactic body radiation therapy and surgery in stage I non-small cell lung cancer. J. Thorac. Oncol. 13, 689–698 10.1016/j.jtho.2018.01.021 - DOI - PubMed
-
- Liu D., Nakashima N., Nakano J., Tarumi S., Matsuura N., Nakano T.. et al. (2017) Customized adjuvant chemotherapy based on biomarker examination may improve survival of patients completely resected for non-small-cell lung cancer. Anticancer Res. 37, 2501–2507 10.21873/anticanres.11591 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

