Controlling aflatoxin contamination and propagation of Aspergillus flavus by a soy-fermenting Aspergillus oryzae strain
- PMID: 30442975
- PMCID: PMC6237848
- DOI: 10.1038/s41598-018-35246-1
Controlling aflatoxin contamination and propagation of Aspergillus flavus by a soy-fermenting Aspergillus oryzae strain
Abstract
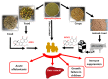
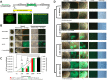
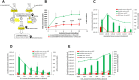
Aflatoxins (AFs) are a group of carcinogenic and immunosuppressive mycotoxins that threaten global food safety. Globally, over 4.5 billion people are exposed to unmonitored levels of AFs. Aspergillus flavus is the major source of AF contamination in agricultural crops. One approach to reduce levels of AFs in agricultural commodities is to apply a non-aflatoxigenic competitor, e.g., Afla-Guard, to crop fields. In this study, we demonstrate that the food fermenting Aspergillus oryzae M2040 strain, isolated from Korean Meju (a brick of dry-fermented soybeans), can inhibit aflatoxin B1 (AFB1) production and proliferation of toxigenic A. flavus in lab culture conditions and peanuts. In peanuts, 1% inoculation level of A. oryzae M2040 could effectively displace the toxigenic A. flavus and inhibit AFB1 production. Moreover, cell-free culture filtrate of A. oryzae M2040 effectively inhibited AFB1 production and A. flavus growth, suggesting A. oryzae M2040 secretes inhibitory compounds. Whole genome-based comparative analyses indicate that the A. oryzae M2040 and Afla-Guard genomes are 37.9 and 36.4 Mbp, respectively, with each genome containing ~100 lineage specific genes. Our study establishes the idea of using A. oryzae and/or its cell-free culture fermentate as a potent biocontrol agent to control A. flavus propagation and AF contamination.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

