Reevaluating the Salty Divide: Phylogenetic Specificity of Transitions between Marine and Freshwater Systems
- PMID: 30443603
- PMCID: PMC6234284
- DOI: 10.1128/mSystems.00232-18
Reevaluating the Salty Divide: Phylogenetic Specificity of Transitions between Marine and Freshwater Systems
Abstract
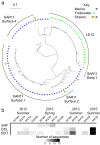
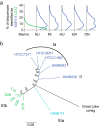
Marine and freshwater microbial communities are phylogenetically distinct, and transitions between habitat types are thought to be infrequent. We compared the phylogenetic diversity of marine and freshwater microorganisms and identified specific lineages exhibiting notably high or low similarity between marine and freshwater ecosystems using a meta-analysis of 16S rRNA gene tag-sequencing data sets. As expected, marine and freshwater microbial communities differed in the relative abundance of major phyla and contained habitat-specific lineages. At the same time, and contrary to expectations, many shared taxa were observed in both habitats. Based on several metrics, we found that Gammaproteobacteria, Alphaproteobacteria, Bacteroidetes, and Betaproteobacteria contained the highest number of closely related marine and freshwater sequences, suggesting comparatively recent habitat transitions in these groups. Using the abundant alphaproteobacterial group SAR11 as an example, we found evidence that new lineages, beyond the recognized LD12 clade, are detected in freshwater at low but reproducible abundances; this evidence extends beyond the 16S rRNA locus to core genes throughout the genome. Our results suggest that shared taxa are numerous, but tend to occur sporadically and at low relative abundance in one habitat type, leading to an underestimation of transition frequency between marine and freshwater habitats. Rare taxa with abundances near or below detection, including lineages that appear to have crossed the salty divide relatively recently, may possess adaptations enabling them to exploit opportunities for niche expansion when environments are disturbed or conditions change. IMPORTANCE The distribution of microbial diversity across environments yields insight into processes that create and maintain this diversity as well as potential to infer how communities will respond to future environmental changes. We integrated data sets from dozens of freshwater lake and marine samples to compare diversity across open water habitats differing in salinity. Our novel combination of sequence-based approaches revealed lineages that likely experienced a recent transition across habitat types. These taxa are promising targets for studying physiological constraints on salinity tolerance. Our findings contribute to understanding the ecological and evolutionary controls on microbial distributions, and open up new questions regarding the plasticity and adaptability of particular lineages.
Keywords: 16S rRNA; SAR11; aquatic ecology; aquatic microbiology; biogeography; environmental transitions; microbial ecology; tag sequencing.
Figures



References
-
- Cole JJ. 1982. Interactions between bacteria and algae in aquatic ecosystems. Annu Rev Ecol Syst 13:291–314. doi: 10.1146/annurev.es.13.110182.001451. - DOI
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
