MiR-130b promotes the progression of oesophageal squamous cell carcinoma by targeting SASH1
- PMID: 30443973
- PMCID: PMC6307769
- DOI: 10.1111/jcmm.13887
MiR-130b promotes the progression of oesophageal squamous cell carcinoma by targeting SASH1
Abstract
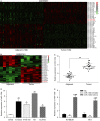
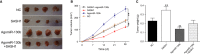
MiR-130b and SAM and SH3 domain containing 1 (SASH1) play an important role in many types of human cancers. The aim of our research was to study their interactions in the process of the proliferation and aggressiveness of oesophageal squamous cell carcinoma (ESCC) cells. Microarray analysis was done to screen the differentially expressed genes in the ESCC tissues. miR-130b and SASH1 mRNA levels in the ESCC tissues and cells were detected by qRT-PCR. Dual luciferase reporter system was used to verify the target relationship between miR-130b and SASH1. The effects of miR-130b on SASH1 expression were explored by western blot in KYSE30 and TE1 cell lines. CCK-8 assay, flow cytometry, Transwell, and wound healing assays were conducted to explore the effects of miR-130b and SASH1 in vitro. In addition, in vivo experiments were conducted to study the roles of miR-130b and SASH1. miR-130b was highly expressed, while SASH1 was the opposite in both the ESCC tissues and cells. The expression of SASH1 was inhibited by the direct binding of miR-130b. The inhibition of miR-130b reduced the proliferation and aggressiveness of ESCC cells, while it also induced apoptosis and cell cycle arrest in the ESCC cells by suppressing SASH1. The in vivo assay suggested that the overexpression of miR-130b promoted the growth of ESCC tumours. MiR-130b was up-regulated in the ESCC tumour tissues and cells, acting as a tumour promoter. A stimulating effect was demonstrated on ESCC cell growth and aggressiveness by suppressing SASH1, which is an anti-oncogene.
Keywords: SASH1; MiR-130b; esophageal squamous cell carcinoma.
© 2018 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures





References
-
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241‐2252. - PubMed
-
- Matsushima K, Isomoto H, Kohno S, et al. MicroRNAs and esophageal squamous cell carcinoma. Digestion. 2010;82:138‐144. - PubMed
-
- Luthra R, Wu TT, Luthra MG, et al. Gene expression profiling of localized esophageal carcinomas: association with pathologic response to preoperative chemoradiation. J Clin Oncol. 2006;24:259‐267. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

