TGF-β1 promotes gap junctions formation in chondrocytes via Smad3/Smad4 signalling
- PMID: 30444057
- PMCID: PMC6495951
- DOI: 10.1111/cpr.12544
TGF-β1 promotes gap junctions formation in chondrocytes via Smad3/Smad4 signalling
Abstract
Objectives: Connexin-mediated functional gap junction intercellular communication (GJIC) has a vital role in development, homeostasis and pathology. Transforming growth factor-β1 (TGF-β1), as one of the most vital factors in chondrocytes, promotes cartilage precursor cell differentiation and chondrocyte proliferation, migration and metabolism. However, how TGF-β1 mediates GJIC in chondrocytes remains unclear. This study aims to determine the influence of TGF-β1 on GJIC in mouse chondrocytes and its underlying mechanism.
Methods: qPCR and mRNA microarray were used to verify the expression of genes in the TGF-β and connexin families in cartilage and chondrocytes. A scrape loading/dye transfer assay was performed to explore GJIC. Western blot analysis was used to detect connexin43 (Cx43) and Smad signalling components. Immunofluorescence staining was performed to characterize protein distribution.
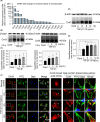
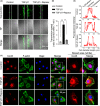
Results: The TGF-β1 mRNA was the highest expressed member of the TGFβ super family in cartilage. TGF-β1 promoted functional GJIC through increased expression of Cx43. TGF-β1-mediated GJIC required the participation of TGF-β type I receptor. TGF-β1 activated Smad3 and Smad4 signalling to facilitate their nuclear translocation. The Smad3 and Smad4 signalling proteins bound to the promoter of Gja1 and thus initiated Cx43 gene expression.
Conclusions: For the first time, these results revealed a vital role of TGF-β1 in cell-cell communication in chondrocytes via gap junction formation. We describe the regulatory mechanism, the involvement of TGF-β type I receptor and the nuclear translocation of Smad3/4.
Keywords: GJIC; Smad3; Smad4; TGF-β1; chondrocyte; connexin43.
© 2018 The Authors. Cell Proliferation Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




Similar articles
-
TGF-β2 increases cell-cell communication in chondrocytes via p-Smad3 signalling.Biochim Biophys Acta Mol Cell Res. 2022 Feb;1869(2):119175. doi: 10.1016/j.bbamcr.2021.119175. Epub 2021 Dec 1. Biochim Biophys Acta Mol Cell Res. 2022. PMID: 34863793
-
The involvement of the ERK-MAPK pathway in TGF-β1-mediated connexin43-gap junction formation in chondrocytes.Connect Tissue Res. 2019 Sep;60(5):477-486. doi: 10.1080/03008207.2019.1593394. Epub 2019 Mar 22. Connect Tissue Res. 2019. PMID: 30897973
-
Transforming growth factor-β1 up-regulates connexin43 expression in human granulosa cells.Hum Reprod. 2015 Sep;30(9):2190-201. doi: 10.1093/humrep/dev175. Epub 2015 Jul 22. Hum Reprod. 2015. PMID: 26202915 Free PMC article.
-
Transforming growth factor-β1 up-regulates connexin43 expression in osteocytes via canonical Smad-dependent signaling pathway.Biosci Rep. 2018 Dec 14;38(6):BSR20181678. doi: 10.1042/BSR20181678. Print 2018 Dec 21. Biosci Rep. 2018. PMID: 30482881 Free PMC article.
-
SMAD4 contributes to chondrocyte and osteocyte development.J Cell Mol Med. 2022 Jan;26(1):1-15. doi: 10.1111/jcmm.17080. Epub 2021 Nov 28. J Cell Mol Med. 2022. PMID: 34841647 Free PMC article. Review.
Cited by
-
The role of TGF-beta3 in cartilage development and osteoarthritis.Bone Res. 2023 Jan 2;11(1):2. doi: 10.1038/s41413-022-00239-4. Bone Res. 2023. PMID: 36588106 Free PMC article. Review.
-
Gap junction-mediated cell-to-cell communication in oral development and oral diseases: a concise review of research progress.Int J Oral Sci. 2020 Jun 12;12(1):17. doi: 10.1038/s41368-020-0086-6. Int J Oral Sci. 2020. PMID: 32532966 Free PMC article. Review.
-
Cryptotanshinone Attenuates Airway Remodeling by Inhibiting Crosstalk Between Tumor Necrosis Factor-Like Weak Inducer of Apoptosis and Transforming Growth Factor Beta 1 Signaling Pathways in Asthma.Front Pharmacol. 2019 Nov 11;10:1338. doi: 10.3389/fphar.2019.01338. eCollection 2019. Front Pharmacol. 2019. PMID: 31780948 Free PMC article.
-
Valproic acid enhances neurosphere formation in cultured rat embryonic cortical cells through TGFβ1 signaling.J Biomed Res. 2022 Feb 15;36(2):127-140. doi: 10.7555/JBR.36.20210109. J Biomed Res. 2022. PMID: 35387900 Free PMC article.
-
Expression and Regulatory Network Analysis of BICC1 for Aged Sca-1-Positive Bone Narrow Mesenchymal Stem Cells.Dis Markers. 2022 Jun 15;2022:4759172. doi: 10.1155/2022/4759172. eCollection 2022. Dis Markers. 2022. PMID: 35756494 Free PMC article.
References
-
- Giepmans B. Gap junctions and connexin‐interacting proteins. Cardiovasc Res. 2004;62:233‐245. - PubMed
-
- Hervé JC, Derangeon M. Gap‐junction‐mediated cell‐to‐cell communication. Cell Tissue Res. 2013;352:21‐31. - PubMed
-
- Willecke K, Eiberger J, Degen J, et al. Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem. 2002;383:725‐737. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

