Genome-wide detection of terpene synthase genes in holy basil (Ocimum sanctum L.)
- PMID: 30444870
- PMCID: PMC6239295
- DOI: 10.1371/journal.pone.0207097
Genome-wide detection of terpene synthase genes in holy basil (Ocimum sanctum L.)
Abstract
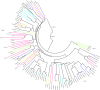
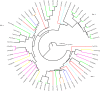
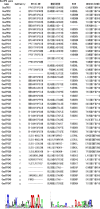
Holy basil (Ocimum sanctum L.) and sweet basil (Ocimum basilicum L.) are the most commonly grown basil species in India for essential oil production and biosynthesis of potentially volatile and non-volatile phytomolecules with commercial significance. The aroma, flavor and pharmaceutical value of Ocimum species is a significance of its essential oil, which contains most of the monoterpenes and sesquiterpenes. A large number of plants have been studied for characterization and identification of terpene synthase genes, involved in terpenoids biosynthesis. The goal of this study is to discover and identify the putative functional terpene synthase genes in O. sanctum. HMMER search was performed by using a set of 13 well sequenced and annotated plant genomes including the newly sequenced genome of O. sanctum with Pfam-A database locally, using HMMER 3.0 hmmsearch for the two Pfam domains (PF01397 and PF03936). Using this search method 81 putative terpene synthases genes (OsaTPS) were identified in O. sanctum; the study further reveals 47 OsaTPS were putatively functional genes, 19 partial OsaTPS, and 15 OsaTPS as probably pseudogenes. All these identified OsaTPS genes were compared with other plant species, and phylogenetic analysis reveals the subfamily classification of OsaTPS in TPS-a, -b, -c, -e, -f and TPS-g subfamilies clusters. This genome-wide identification of OsaTPS genes, their phylogenetic analysis and secondary metabolite pathway mapping predictions together provide a comprehensive understanding of the TPS gene family in Ocimum sanctum and offer opportunities for the characterization and functional validation of numbers of terpene synthase genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Holstein SA, Hohl RJ. Isoprenoids: remarkable diversity of form and function. Lipids. 2004;39: 293–309. Available: http://www.ncbi.nlm.nih.gov/pubmed/15357017 - PubMed
-
- Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep. 1999;16: 565–74. Available: http://www.ncbi.nlm.nih.gov/pubmed/10584331 - PubMed
-
- Lichtenthaler HK. THE 1-DEOXY-D-XYLULOSE-5-PHOSPHATE PATHWAY OF ISOPRENOID BIOSYNTHESIS IN PLANTS. Annu Rev Plant Physiol Plant Mol Biol. 1999;50: 47–65. 10.1146/annurev.arplant.50.1.47 - DOI - PubMed
-
- Sapir-Mir M, Mett A, … EB-P, 2008. undefined. Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is. Am Soc Plant Biol. Available: http://www.plantphysiol.org/content/148/3/1219.short - PMC - PubMed
-
- Külheim C, Yeoh SH, Wallis IR, Laffan S, Moran GF, Foley WJ. The molecular basis of quantitative variation in foliar secondary metabolites in Eucalyptus globulus. New Phytol. 2011;191: 1041–53. 10.1111/j.1469-8137.2011.03769.x - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

