Functional regulatory mechanism of smooth muscle cell-restricted LMOD1 coronary artery disease locus
- PMID: 30444878
- PMCID: PMC6268002
- DOI: 10.1371/journal.pgen.1007755
Functional regulatory mechanism of smooth muscle cell-restricted LMOD1 coronary artery disease locus
Abstract
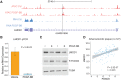
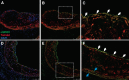
Recent genome-wide association studies (GWAS) have identified multiple new loci which appear to alter coronary artery disease (CAD) risk via arterial wall-specific mechanisms. One of the annotated genes encodes LMOD1 (Leiomodin 1), a member of the actin filament nucleator family that is highly enriched in smooth muscle-containing tissues such as the artery wall. However, it is still unknown whether LMOD1 is the causal gene at this locus and also how the associated variants alter LMOD1 expression/function and CAD risk. Using epigenomic profiling we recently identified a non-coding regulatory variant, rs34091558, which is in tight linkage disequilibrium (LD) with the lead CAD GWAS variant, rs2820315. Herein we demonstrate through expression quantitative trait loci (eQTL) and statistical fine-mapping in GTEx, STARNET, and human coronary artery smooth muscle cell (HCASMC) datasets, rs34091558 is the top regulatory variant for LMOD1 in vascular tissues. Position weight matrix (PWM) analyses identify the protective allele rs34091558-TA to form a conserved Forkhead box O3 (FOXO3) binding motif, which is disrupted by the risk allele rs34091558-A. FOXO3 chromatin immunoprecipitation and reporter assays show reduced FOXO3 binding and LMOD1 transcriptional activity by the risk allele, consistent with effects of FOXO3 downregulation on LMOD1. LMOD1 knockdown results in increased proliferation and migration and decreased cell contraction in HCASMC, and immunostaining in atherosclerotic lesions in the SMC lineage tracing reporter mouse support a key role for LMOD1 in maintaining the differentiated SMC phenotype. These results provide compelling functional evidence that genetic variation is associated with dysregulated LMOD1 expression/function in SMCs, together contributing to the heritable risk for CAD.
Conflict of interest statement
JLMB is a founder, consultant and major shareholder of Clinical Gene Networks AB (CGN) together with AR. OF is a part-time employee of CGN. JLMB, AR and EES are members of the board of directors. CGN has an invested interest in the STARNET biobank and data set. However, CGN has expressed no claims or sought any patents related to the results presented in this manuscript.
Figures





References
-
- CARDIoGRAMplusC4D Consortium, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45: 25–33. 10.1038/ng.2480 - DOI - PMC - PubMed
-
- Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. Nature Publishing Group; 2011;43: 339–344. 10.1038/ng.782 - DOI - PubMed
-
- Nikpay M, Goel A, Won H-H, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47: 1121–1130. 10.1038/ng.3396 - DOI - PMC - PubMed
-
- Howson JMM, Zhao W, Barnes DR, Ho W-K, Young R, Paul DS, et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet. 2017;49: 1113–1119. 10.1038/ng.3874 - DOI - PMC - PubMed
-
- Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49: 1385–1391. 10.1038/ng.3913 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99 HL136865/HL/NHLBI NIH HHS/United States
- R01 HL125863/HL/NHLBI NIH HHS/United States
- R00 HL136865/HL/NHLBI NIH HHS/United States
- R01 HG008150/HG/NHGRI NIH HHS/United States
- K99 HL125912/HL/NHLBI NIH HHS/United States
- R01 HL125224/HL/NHLBI NIH HHS/United States
- R00 HL125912/HL/NHLBI NIH HHS/United States
- R01 HL071207/HL/NHLBI NIH HHS/United States
- R01 HL123370/HL/NHLBI NIH HHS/United States
- P30 DK116074/DK/NIDDK NIH HHS/United States
- R33 HL120757/HL/NHLBI NIH HHS/United States
- R21 HL120757/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

