The Effect of Influenza Virus on the Human Oropharyngeal Microbiome
- PMID: 30445563
- PMCID: PMC6541733
- DOI: 10.1093/cid/ciy821
The Effect of Influenza Virus on the Human Oropharyngeal Microbiome
Abstract
Background: Secondary bacterial infections are an important cause of morbidity and mortality associated with influenza infections. As bacterial disease can be caused by a disturbance of the host microbiome, we examined the impact of influenza on the upper respiratory tract microbiome in a human challenge study.
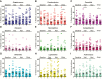
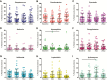
Methods: The dynamics and ecology of the throat microbiome were examined following an experimental influenza challenge of 52 previously-healthy adult volunteers with influenza A/Wisconsin/67/2005 (H3N2) by intranasal inoculation; 35 healthy control subjects were not subjected to the viral challenge. Serial oropharyngeal samples were taken over a 30-day period, and the V1-V3 region of the bacterial 16S ribosomal RNA sequences were amplified and sequenced to determine the composition of the microbiome. The carriage of pathogens was also detected.
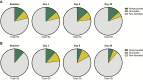
Results: Of the 52 challenged individuals, 43 developed proven influenza infections, 33 of whom became symptomatic. None of the controls developed influenza, although 22% reported symptoms. The diversity of bacterial communities remained remarkably stable following the acquisition of influenza, with no significant differences over time between individuals with influenza and those in the control group. Influenza infection was not associated with perturbation of the microbiome at the level of phylum or genus. There was no change in colonization rates with Streptococcus pneumoniae or Neisseria meningitidis.
Conclusions: The throat microbiota is resilient to influenza infection, indicating the robustness of the upper-airway microbiome.
Keywords: influenza; microbiome; upper respiratory tract.
© The Author(s) 2018. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures






References
-
- Morens DM, Fauci AS. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis 2007; 195:1018–28. - PubMed
-
- Lee EH, Wu C, Lee EU, et al. . Fatalities associated with the 2009 H1N1 influenza A virus in New York city. Clin Infect Dis 2010; 50:1498–504. - PubMed
-
- Smith W, Andrewes CH, Laidlaw PP. A virus obtained from influenza patients. Lancet 1933; 2:66–8.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

