pyNVR: investigating factors affecting feature selection from scRNA-seq data for lineage reconstruction
- PMID: 30445607
- PMCID: PMC6596893
- DOI: 10.1093/bioinformatics/bty950
pyNVR: investigating factors affecting feature selection from scRNA-seq data for lineage reconstruction
Abstract
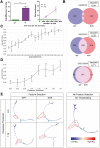
Motivation: The emergence of single-cell RNA-sequencing has enabled analyses that leverage transitioning cell states to reconstruct pseudotemporal trajectories. Multidimensional data sparsity, zero inflation and technical variation necessitate the selection of high-quality features that feed downstream analyses. Despite the development of numerous algorithms for the unsupervised selection of biologically relevant features, their differential performance remains largely unaddressed.
Results: We implemented the neighborhood variance ratio (NVR) feature selection approach as a Python package with substantial improvements in performance. In comparing NVR with multiple unsupervised algorithms such as dpFeature, we observed striking differences in features selected. We present evidence that quantifiable dataset properties have observable and predictable effects on the performance of these algorithms.
Availability and implementation: pyNVR is freely available at https://github.com/KenLauLab/NVR.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2018. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Elowitz M.B. (2002) Stochastic gene expression in a single cell. Science, 297, 1183–1186. - PubMed

