Lateral Flow Immunoassay for Rapid Detection of Grapevine Leafroll-Associated Virus
- PMID: 30445781
- PMCID: PMC6315891
- DOI: 10.3390/bios8040111
Lateral Flow Immunoassay for Rapid Detection of Grapevine Leafroll-Associated Virus
Abstract
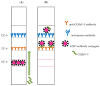
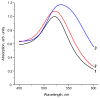
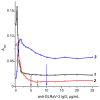
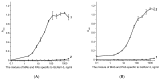
Grapevine leafroll-associated virus 3 (GLRaV-3) is one of the main pathogens of grapes, causing a significant loss in yield and decrease in quality for this agricultural plant. For efficient widespread control of this infection, rapid and simple analytical techniques of on-site testing are requested as a complementary addition for the currently applied hybridization (PCR) and immunoenzyme (ELISA) approaches. The given paper presents development and approbation of the immunochromatographic assay (ICA) for rapid detection of GLRaV-3. The ICA realizes a sandwich immunoassay format with the obtaining complexes ((antibody immobilized on immunochromatographic membrane)⁻(virus in the sample)⁻(antibody immobilized on gold nanoparticles (GNP)) during sample flow along the membrane compounds of the test strip. Three preparations of GNPs were compared for detection of GLRaV-3 at different dilutions of virus-containing sample. The GNPs with maximal average diameters of 51.0 ± 7.9 nm provide GLRaV-3 detection for its maximal dilutions, being 4 times more than when using GNPs with a diameter of 28.3 ± 3.3 nm, and 8 times more than when using GNPs with a diameter of 18.5 ± 3.3 nm. Test strips have been manufactured using the largest GNPs conjugated with anti-GLRaV-3 antibodies at a ratio of 1070:1. When testing samples containing other grape wine viruses, the test strips have not demonstrated staining in the test zone, which confirms the ICA specificity. The approbation of the manufactured test strips indicated that when using ELISA as a reference method, the developed ICA is characterized by a sensitivity of 100% and a specificity of 92%. If PCR is considered as a reference method, then the sensitivity of ICA is 93% and the specificity is 92%. The proposed ICA can be implemented in one stage without the use of any additional reactants or devices. The testing results can be obtained in 10 min and detected visually. It provides significant improvement in GLRaV-3 detection, and the presented approach can be transferred for the development of test systems for other grape wine pathogens.
Keywords: agricultural control; gold nanoparticles; immunochromatography; on-site testing; phytopathogens; test strips.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Maliogka V.I., Martelli G.P., Fuchs M., Katis N.I. Control of viruses infecting grapevine. In: Loebenstein G., Katis N.I., editors. Advance in Virus Research, Control of Plant Virus Diseases Vegetatively-Propagated Crops. Volume 91. Academic Press; Boston, MA, USA: 2015. pp. 175–227. - PubMed
-
- Meng B., Martelli G.P., Golino D.A., Fuchs M., editors. Grapevine Viruses: Molecular Biology, Diagnostics and Management. Springer; Heidelberg, Germany: 2017.
-
- Burger J.T., Maree H.J., Gouveia P., Naidu R.A. Grapevine leafroll-associated virus 3. In: Meng B., Martelli G.P., Golino D.A., Fuchs M., editors. Grapevine Viruses: Molecular Biology, Diagnostics and Management. Springer; Heidelberg, Germany: 2017. pp. 167–196.
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

