Global Diversity of the Brachypodium Species Complex as a Resource for Genome-Wide Association Studies Demonstrated for Agronomic Traits in Response to Climate
- PMID: 30446522
- PMCID: PMC6325704
- DOI: 10.1534/genetics.118.301589
Global Diversity of the Brachypodium Species Complex as a Resource for Genome-Wide Association Studies Demonstrated for Agronomic Traits in Response to Climate
Abstract
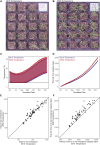
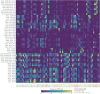
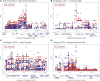
The development of model systems requires a detailed assessment of standing genetic variation across natural populations. The Brachypodium species complex has been promoted as a plant model for grass genomics with translation to small grain and biomass crops. To capture the genetic diversity within this species complex, thousands of Brachypodium accessions from around the globe were collected and genotyped by sequencing. Overall, 1897 samples were classified into two diploid or allopolyploid species, and then further grouped into distinct inbred genotypes. A core set of diverse B. distachyon diploid lines was selected for whole genome sequencing and high resolution phenotyping. Genome-wide association studies across simulated seasonal environments was used to identify candidate genes and pathways tied to key life history and agronomic traits under current and future climatic conditions. A total of 8, 22, and 47 QTL were identified for flowering time, early vigor, and energy traits, respectively. The results highlight the genomic structure of the Brachypodium species complex, and the diploid lines provided a resource that allows complex trait dissection within this grass model species.
Keywords: Brachypodium distachyon; agronomic traits; climate change; climate simulation; ecogenomics; genome-wide association studies; genotyping; plant physiology; population genetics.
Copyright © 2019 Wilson et al.
Figures

References
-
- Bettgenhaeuser J., Corke F. M. K., Opanowicz M., Green P., Hernández-Pinzón I., et al. , 2017. Natural variation in Brachypodium links vernalization and flowering time loci as major flowering determinants. Plant Physiol. 173: 256–268. Available at: http://www.plantphysiol.org/lookup/doi/10.1104/pp.16.00813. 10.1104/pp.16.00813 - DOI - DOI - PMC - PubMed

