RNA Sequencing-Based Transcriptional Overview of Xerotolerance in Cronobacter sakazakii SP291
- PMID: 30446557
- PMCID: PMC6344630
- DOI: 10.1128/AEM.01993-18
RNA Sequencing-Based Transcriptional Overview of Xerotolerance in Cronobacter sakazakii SP291
Abstract
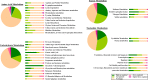
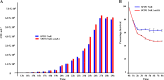
Cronobacter sakazakii is a xerotolerant neonatal pathogen epidemiologically linked to powdered infant food formula, often resulting in high mortality rates. Here, we used transcriptome sequencing (RNA-seq) to provide transcriptional insights into the survival of C. sakazakii in desiccated conditions. Our RNA-seq data show that about 22% of the total C. sakazakii genes were significantly upregulated and 9% were downregulated during desiccation survival. When reverse transcription-quantitative PCR (qRT-PCR) was used to validate the RNA-seq data, we found that the primary desiccation response was gradually downregulated during the tested 4 hours of desiccation, while the secondary response remained constitutively upregulated. The 4-hour desiccation tolerance of C. sakazakii was dependent on the immediate microenvironment surrounding the bacterial cell. The removal of Trypticase soy broth (TSB) salts and the introduction of sterile infant formula residues in the microenvironment enhanced the desiccation survival of C. sakazakii SP291. The trehalose biosynthetic pathway encoded by otsA and otsB, a prominent secondary bacterial desiccation response, was highly upregulated in desiccated C. sakazakiiC. sakazakii SP291 ΔotsAB was significantly inhibited compared with the isogenic wild type in an 8-hour desiccation survival assay, confirming the physiological importance of trehalose in desiccation survival. Overall, we provide a comprehensive RNA-seq-based transcriptional overview along with confirmation of the phenotypic importance of trehalose metabolism in Cronobacter sakazakii during desiccation.IMPORTANCECronobacter sakazakii is a pathogen of importance to neonatal health and is known to persist in dry food matrices, such as powdered infant formula (PIF) and its associated production environment. When infections are reported in neonates, mortality rates can be high. The success of this bacterium in surviving these low-moisture environments suggests that Cronobacter species can respond to a variety of environmental signals. Therefore, understanding those signals that aid the persistence of this pathogen in these ecological niches is an important step toward the development of strategies to reduce the risk of contamination of PIF. This research led to the identification of candidate genes that play a role in the persistence of this pathogen in desiccated conditions and, thereby, serve as a model target to design future strategies to mitigate PIF-associated survival of C. sakazakii.
Keywords: Cronobacter sakazakii; RNA-seq; desiccation; transcriptome; xerotolerance.
Copyright © 2019 American Society for Microbiology.
Figures






References
-
- Lund BM, Baird-Parker TC, Gould GW. 2000. The microbiological safety and quality of food. Aspen Publishers, Gaithersburg, MD.
-
- Carrasco E, Morales-Rueda A, García-Gimeno RM. 2012. Cross-contamination and recontamination by Salmonella in foods: a review. Food Res Int 45:545–556. doi:10.1016/j.foodres.2011.11.004. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

