Novel Cholera Toxin Variant and ToxT Regulon in Environmental Vibrio mimicus Isolates: Potential Resources for the Evolution of Vibrio cholerae Hybrid Strains
- PMID: 30446560
- PMCID: PMC6344616
- DOI: 10.1128/AEM.01977-18
Novel Cholera Toxin Variant and ToxT Regulon in Environmental Vibrio mimicus Isolates: Potential Resources for the Evolution of Vibrio cholerae Hybrid Strains
Abstract
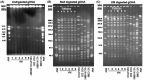
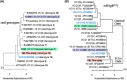
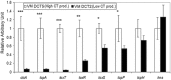
Atypical El Tor strains of Vibrio cholerae O1 harboring variant ctxB genes of cholera toxin (CT) have gradually become a major cause of recent cholera epidemics. Vibrio mimicus occasionally produces CT, encoded by ctxAB on CTXФ genome; toxin-coregulated pilus (TCP), a major intestinal colonization factor; and also the CTXФ-specific receptor. This study carried out extensive molecular characterization of CTXФ and ToxT regulon in V. mimicusctx-positive (ctx+) strains (i.e., V. mimicus strains containing ctx) isolated from the Bengal coast. Southern hybridization, PCR, and DNA sequencing of virulence-related genes revealed the presence of an El Tor type CTX prophage (CTXET) carrying a novel ctxAB, tandem copies of environmental type pre-CTX prophage (pre-CTXEnv), and RS1 elements, which were organized as an RS1-CTXET-RS1-pre-CTXEnv-pre-CTXEnv array. Additionally, novel variants of tcpA and toxT, respectively, showing phylogenetic lineage to a clade of V. cholerae non-O1 and to a clade of V. cholerae non-O139, were identified. The V. mimicus strains lacked the RTX (repeat in toxin) and TLC (toxin-linked cryptic) elements and lacked Vibrio seventh-pandemic islands of the El Tor strains but contained five heptamer (TTTTGAT) repeats in ctxAB promoter region similar to those seen with some classical strains of V. cholerae O1. Pulsed-field gel electrophoresis (PFGE) analysis showed that all the ctx+V. mimicus strains were clonally related. However, their in vitro CT production and in vivo toxigenicity characteristics were variable, which could be explainable by differential transcription of virulence genes along with the ToxR regulon. Taken together, our findings strongly suggest that environmental V. mimicus strains act as a potential reservoir of atypical virulence factors, including variant CT and ToxT regulons, and may contribute to the evolution of V. cholerae hybrid strains.IMPORTANCE Natural diversification of CTXФ and ctxAB genes certainly influences disease severity and shifting patterns in major etiological agents of cholera, e.g., the overwhelming emergence of hybrid El Tor variants, replacing the prototype El Tor strains of V. cholerae This report, showing the occurrence of CTXET comprising a novel variant of ctxAB in V. mimicus, points out a previously unnoticed evolutionary event that is independent of the evolutionary event associated with the El Tor strains of V. cholerae Identification and cluster analysis of the newly discovered alleles of tcpA and toxT suggest their horizontal transfer from an uncommon clone of V. cholerae The genomic contents of ToxT regulon and of tandemly arranged multiple pre-CTXФEnv and of a CTXФET in V. mimicus probably act as salient raw materials that induce natural recombination among the hallmark virulence genes of hybrid V. cholerae strains. This report provides valuable information to enrich our knowledge on the evolution of new variant CT and ToxT regulons.
Keywords: CTXФ; El Tor biotype; Vibrio cholerae; Vibrio mimicus; cholera toxin; classical biotype; tcpA; toxT.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Rashed SM, Mannan SB, Johura FT, Islam MT, Sadique A, Watanabe H, Sack RB, Huq A, Colwell RR, Cravioto A, Alam M. 2012. Genetic characteristics of drug-resistant Vibrio cholerae O1 causing endemic cholera in Dhaka, 2006–2011. J Med Microbiol 61:1736–1745. doi:10.1099/jmm.0.049635-0. - DOI - PMC - PubMed
-
- Adewale AK, Pazhani GP, Abiodun IB, Afolabi O, Kolawole OD, Mukhopadhyay AK, Ramamurthy T. 2016. Unique clones of Vibrio cholerae O1 El Tor with Haitian type ctxB allele implicated in the recent cholera epidemics from Nigeria, Africa. PLoS One 11:e0159794. doi:10.1371/journal.pone.0159794. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

