A majority of HIV persistence during antiretroviral therapy is due to infected cell proliferation
- PMID: 30446650
- PMCID: PMC6240116
- DOI: 10.1038/s41467-018-06843-5
A majority of HIV persistence during antiretroviral therapy is due to infected cell proliferation
Abstract
Antiretroviral therapy (ART) suppresses viral replication in people living with HIV. Yet, infected cells persist for decades on ART and viremia returns if ART is stopped. Persistence has been attributed to viral replication in an ART sanctuary and long-lived and/or proliferating latently infected cells. Using ecological methods and existing data, we infer that >99% of infected cells are members of clonal populations after one year of ART. We reconcile our results with observations from the first months of ART, demonstrating mathematically how a fossil record of historic HIV replication permits observed viral evolution even while most new infected cells arise from proliferation. Together, our results imply cellular proliferation generates a majority of infected cells during ART. Therefore, reducing proliferation could decrease the size of the HIV reservoir and help achieve a functional cure.
Conflict of interest statement
The authors declare no competing interests.
Figures



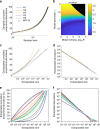
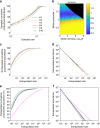



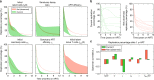
References
Publication types
MeSH terms
Substances
Grants and funding
- K25 AI155224/AI/NIAID NIH HHS/United States
- R01 AI121129/AI/NIAID NIH HHS/United States
- P01 AI030371-24, U19 AI113173-02, R01 AI121129-01, UM1 AI126623, UM1 AI068635/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)/International
- U19 AI096109/AI/NIAID NIH HHS/United States
- T32 AI007044/AI/NIAID NIH HHS/United States
- U19 AI113173/AI/NIAID NIH HHS/United States
- Postdoctoral Fellowship/Washington Research Foundation (WRF)/International
- UM1 AI126611/AI/NIAID NIH HHS/United States
- UM1 AI126623/AI/NIAID NIH HHS/United States
- UM1 AI068635/AI/NIAID NIH HHS/United States
- KL2 TR002317/TR/NCATS NIH HHS/United States
- P30 AI027757/AI/NIAID NIH HHS/United States
- TL1 TR002318/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

