Bardet-Biedl Syndrome proteins regulate cilia disassembly during tissue maturation
- PMID: 30446775
- PMCID: PMC11105770
- DOI: 10.1007/s00018-018-2966-x
Bardet-Biedl Syndrome proteins regulate cilia disassembly during tissue maturation
Abstract
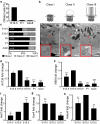
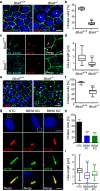
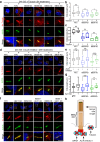
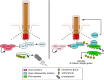
Primary cilia are conserved organelles that mediate cellular communication crucial for organogenesis and homeostasis in numerous tissues. The retinal pigment epithelium (RPE) is a ciliated monolayer in the eye that borders the retina and is vital for visual function. Maturation of the RPE is absolutely critical for visual function and the role of the primary cilium in this process has been largely ignored to date. We show that primary cilia are transiently present during RPE development and that as the RPE matures, primary cilia retract, and gene expression of ciliary disassembly components decline. We observe that ciliary-associated BBS proteins protect against HDAC6-mediated ciliary disassembly via their recruitment of Inversin to the base of the primary cilium. Inhibition of ciliary disassembly components was able to rescue ciliary length defects in BBS deficient cells. This consequently affects ciliary regulation of Wnt signaling. Our results shed light onto the mechanisms by which cilia-mediated signaling facilitates tissue maturation.
Keywords: Ciliopathy; Proteasomal degradation; Retinal dystrophy; Signaling inhibitors; Signaling pathways.
Conflict of interest statement
The authors declares that they have no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

